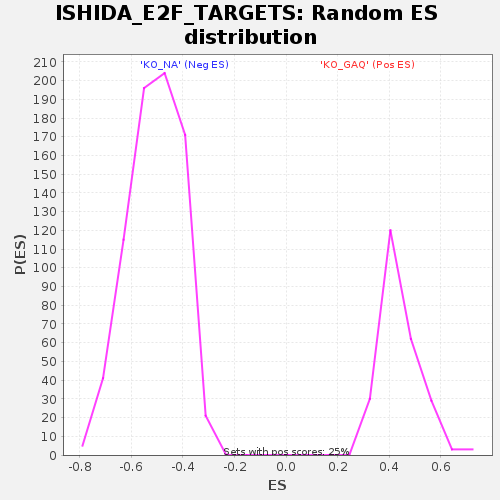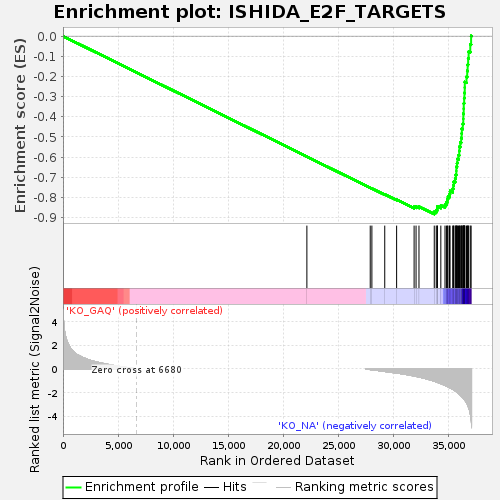
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | ISHIDA_E2F_TARGETS |
| Enrichment Score (ES) | -0.88287574 |
| Normalized Enrichment Score (NES) | -1.7317346 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.1736576E-4 |
| FWER p-Value | 0.005 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ANXA8L1 | annexin A8 like 1 [Source:HGNC Symbol;Acc:HGNC:23334] | 22139 | 0.000 | -0.5977 | No |
| 2 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27890 | -0.049 | -0.7524 | No |
| 3 | DCTPP1 | dCTP pyrophosphatase 1 [Source:HGNC Symbol;Acc:HGNC:28777] | 28040 | -0.066 | -0.7557 | No |
| 4 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 29208 | -0.205 | -0.7851 | No |
| 5 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 30286 | -0.341 | -0.8106 | No |
| 6 | KPNA2 | karyopherin subunit alpha 2 [Source:HGNC Symbol;Acc:HGNC:6395] | 31873 | -0.586 | -0.8472 | No |
| 7 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 32050 | -0.618 | -0.8454 | No |
| 8 | DNA2 | DNA replication helicase/nuclease 2 [Source:HGNC Symbol;Acc:HGNC:2939] | 32315 | -0.677 | -0.8454 | No |
| 9 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33703 | -1.035 | -0.8720 | Yes |
| 10 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33881 | -1.088 | -0.8652 | Yes |
| 11 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 33967 | -1.118 | -0.8557 | Yes |
| 12 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33988 | -1.124 | -0.8444 | Yes |
| 13 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 34299 | -1.231 | -0.8398 | Yes |
| 14 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34678 | -1.382 | -0.8354 | Yes |
| 15 | SGO1 | shugoshin 1 [Source:HGNC Symbol;Acc:HGNC:25088] | 34809 | -1.440 | -0.8237 | Yes |
| 16 | CDT1 | chromatin licensing and DNA replication factor 1 [Source:HGNC Symbol;Acc:HGNC:24576] | 34893 | -1.473 | -0.8104 | Yes |
| 17 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34948 | -1.490 | -0.7962 | Yes |
| 18 | CDCA7 | cell division cycle associated 7 [Source:HGNC Symbol;Acc:HGNC:14628] | 35087 | -1.556 | -0.7835 | Yes |
| 19 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 35135 | -1.581 | -0.7680 | Yes |
| 20 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 35380 | -1.712 | -0.7566 | Yes |
| 21 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 35438 | -1.743 | -0.7397 | Yes |
| 22 | CDK2 | cyclin dependent kinase 2 [Source:HGNC Symbol;Acc:HGNC:1771] | 35470 | -1.760 | -0.7220 | Yes |
| 23 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35602 | -1.838 | -0.7061 | Yes |
| 24 | ASF1B | anti-silencing function 1B histone chaperone [Source:HGNC Symbol;Acc:HGNC:20996] | 35653 | -1.875 | -0.6877 | Yes |
| 25 | DBF4 | DBF4 zinc finger [Source:HGNC Symbol;Acc:HGNC:17364] | 35707 | -1.910 | -0.6690 | Yes |
| 26 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 35709 | -1.910 | -0.6488 | Yes |
| 27 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35769 | -1.949 | -0.6298 | Yes |
| 28 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 35806 | -1.978 | -0.6099 | Yes |
| 29 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35905 | -2.049 | -0.5910 | Yes |
| 30 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35971 | -2.105 | -0.5705 | Yes |
| 31 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 35988 | -2.120 | -0.5486 | Yes |
| 32 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36065 | -2.187 | -0.5275 | Yes |
| 33 | UBE2T | ubiquitin conjugating enzyme E2 T [Source:HGNC Symbol;Acc:HGNC:25009] | 36145 | -2.264 | -0.5058 | Yes |
| 34 | ANLN | anillin actin binding protein [Source:HGNC Symbol;Acc:HGNC:14082] | 36192 | -2.309 | -0.4826 | Yes |
| 35 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36203 | -2.318 | -0.4585 | Yes |
| 36 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36291 | -2.397 | -0.4355 | Yes |
| 37 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36338 | -2.450 | -0.4109 | Yes |
| 38 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 36341 | -2.453 | -0.3851 | Yes |
| 39 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36377 | -2.484 | -0.3598 | Yes |
| 40 | HMGB3 | high mobility group box 3 [Source:HGNC Symbol;Acc:HGNC:5004] | 36380 | -2.489 | -0.3336 | Yes |
| 41 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36418 | -2.536 | -0.3078 | Yes |
| 42 | RPA1 | replication protein A1 [Source:HGNC Symbol;Acc:HGNC:10289] | 36442 | -2.562 | -0.2814 | Yes |
| 43 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36471 | -2.598 | -0.2547 | Yes |
| 44 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36474 | -2.605 | -0.2273 | Yes |
| 45 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 36628 | -2.819 | -0.2017 | Yes |
| 46 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36694 | -2.936 | -0.1725 | Yes |
| 47 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36733 | -3.017 | -0.1416 | Yes |
| 48 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36779 | -3.125 | -0.1099 | Yes |
| 49 | CCNE1 | cyclin E1 [Source:HGNC Symbol;Acc:HGNC:1589] | 36823 | -3.251 | -0.0767 | Yes |
| 50 | RB1 | RB transcriptional corepressor 1 [Source:HGNC Symbol;Acc:HGNC:9884] | 36972 | -3.766 | -0.0410 | Yes |
| 51 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37048 | -4.181 | 0.0011 | Yes |