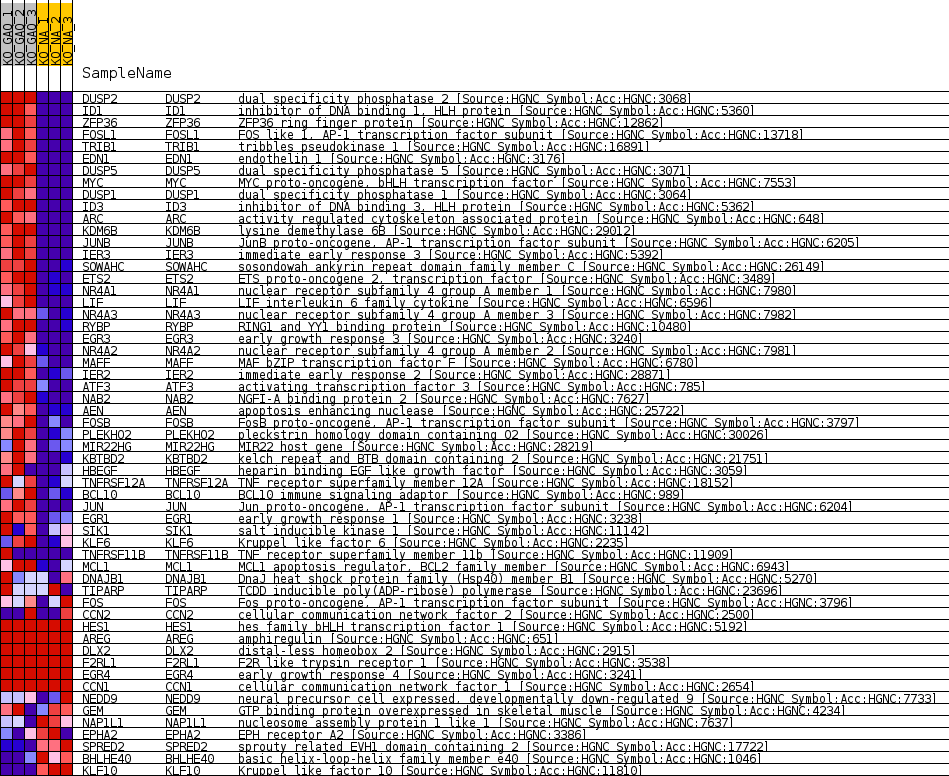
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
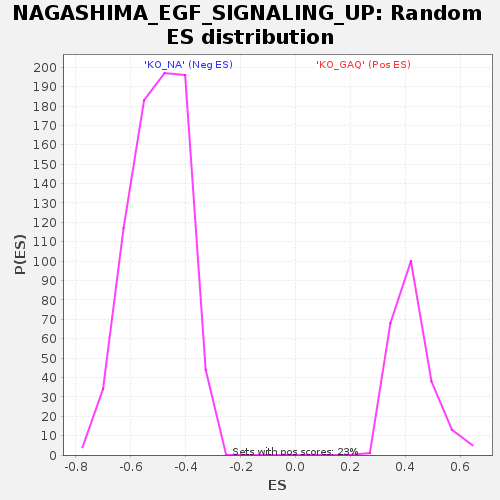
| GeneSet | NAGASHIMA_EGF_SIGNALING_UP |
| Enrichment Score (ES) | 0.8375568 |
| Normalized Enrichment Score (NES) | 1.9789491 |
| Nominal p-value | 0.0 |
| FDR q-value | 1.5430995E-4 |
| FWER p-Value | 0.001 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DUSP2 | dual specificity phosphatase 2 [Source:HGNC Symbol;Acc:HGNC:3068] | 15 | 4.682 | 0.0451 | Yes |
| 2 | ID1 | inhibitor of DNA binding 1, HLH protein [Source:HGNC Symbol;Acc:HGNC:5360] | 24 | 4.487 | 0.0885 | Yes |
| 3 | ZFP36 | ZFP36 ring finger protein [Source:HGNC Symbol;Acc:HGNC:12862] | 27 | 4.450 | 0.1317 | Yes |
| 4 | FOSL1 | FOS like 1, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:13718] | 37 | 4.309 | 0.1734 | Yes |
| 5 | TRIB1 | tribbles pseudokinase 1 [Source:HGNC Symbol;Acc:HGNC:16891] | 38 | 4.281 | 0.2150 | Yes |
| 6 | EDN1 | endothelin 1 [Source:HGNC Symbol;Acc:HGNC:3176] | 44 | 4.198 | 0.2557 | Yes |
| 7 | DUSP5 | dual specificity phosphatase 5 [Source:HGNC Symbol;Acc:HGNC:3071] | 45 | 4.193 | 0.2965 | Yes |
| 8 | MYC | MYC proto-oncogene, bHLH transcription factor [Source:HGNC Symbol;Acc:HGNC:7553] | 47 | 4.168 | 0.3370 | Yes |
| 9 | DUSP1 | dual specificity phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:3064] | 61 | 3.986 | 0.3754 | Yes |
| 10 | ID3 | inhibitor of DNA binding 3, HLH protein [Source:HGNC Symbol;Acc:HGNC:5362] | 71 | 3.870 | 0.4128 | Yes |
| 11 | ARC | activity regulated cytoskeleton associated protein [Source:HGNC Symbol;Acc:HGNC:648] | 78 | 3.741 | 0.4490 | Yes |
| 12 | KDM6B | lysine demethylase 6B [Source:HGNC Symbol;Acc:HGNC:29012] | 92 | 3.598 | 0.4836 | Yes |
| 13 | JUNB | JunB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6205] | 109 | 3.467 | 0.5169 | Yes |
| 14 | IER3 | immediate early response 3 [Source:HGNC Symbol;Acc:HGNC:5392] | 114 | 3.436 | 0.5502 | Yes |
| 15 | SOWAHC | sosondowah ankyrin repeat domain family member C [Source:HGNC Symbol;Acc:HGNC:26149] | 195 | 2.937 | 0.5766 | Yes |
| 16 | ETS2 | ETS proto-oncogene 2, transcription factor [Source:HGNC Symbol;Acc:HGNC:3489] | 230 | 2.775 | 0.6027 | Yes |
| 17 | NR4A1 | nuclear receptor subfamily 4 group A member 1 [Source:HGNC Symbol;Acc:HGNC:7980] | 266 | 2.634 | 0.6273 | Yes |
| 18 | LIF | LIF interleukin 6 family cytokine [Source:HGNC Symbol;Acc:HGNC:6596] | 284 | 2.578 | 0.6519 | Yes |
| 19 | NR4A3 | nuclear receptor subfamily 4 group A member 3 [Source:HGNC Symbol;Acc:HGNC:7982] | 302 | 2.508 | 0.6759 | Yes |
| 20 | RYBP | RING1 and YY1 binding protein [Source:HGNC Symbol;Acc:HGNC:10480] | 370 | 2.328 | 0.6967 | Yes |
| 21 | EGR3 | early growth response 3 [Source:HGNC Symbol;Acc:HGNC:3240] | 467 | 2.108 | 0.7146 | Yes |
| 22 | NR4A2 | nuclear receptor subfamily 4 group A member 2 [Source:HGNC Symbol;Acc:HGNC:7981] | 534 | 1.992 | 0.7322 | Yes |
| 23 | MAFF | MAF bZIP transcription factor F [Source:HGNC Symbol;Acc:HGNC:6780] | 544 | 1.978 | 0.7512 | Yes |
| 24 | IER2 | immediate early response 2 [Source:HGNC Symbol;Acc:HGNC:28871] | 563 | 1.945 | 0.7696 | Yes |
| 25 | ATF3 | activating transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:785] | 582 | 1.916 | 0.7877 | Yes |
| 26 | NAB2 | NGFI-A binding protein 2 [Source:HGNC Symbol;Acc:HGNC:7627] | 594 | 1.884 | 0.8058 | Yes |
| 27 | AEN | apoptosis enhancing nuclease [Source:HGNC Symbol;Acc:HGNC:25722] | 611 | 1.860 | 0.8234 | Yes |
| 28 | FOSB | FosB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3797] | 705 | 1.713 | 0.8376 | Yes |
| 29 | PLEKHO2 | pleckstrin homology domain containing O2 [Source:HGNC Symbol;Acc:HGNC:30026] | 1788 | 0.967 | 0.8177 | No |
| 30 | MIR22HG | MIR22 host gene [Source:HGNC Symbol;Acc:HGNC:28219] | 2063 | 0.868 | 0.8188 | No |
| 31 | KBTBD2 | kelch repeat and BTB domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21751] | 2206 | 0.820 | 0.8229 | No |
| 32 | HBEGF | heparin binding EGF like growth factor [Source:HGNC Symbol;Acc:HGNC:3059] | 2465 | 0.732 | 0.8231 | No |
| 33 | TNFRSF12A | TNF receptor superfamily member 12A [Source:HGNC Symbol;Acc:HGNC:18152] | 2678 | 0.671 | 0.8239 | No |
| 34 | BCL10 | BCL10 immune signaling adaptor [Source:HGNC Symbol;Acc:HGNC:989] | 2697 | 0.665 | 0.8299 | No |
| 35 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6204] | 2706 | 0.664 | 0.8361 | No |
| 36 | EGR1 | early growth response 1 [Source:HGNC Symbol;Acc:HGNC:3238] | 2983 | 0.590 | 0.8344 | No |
| 37 | SIK1 | salt inducible kinase 1 [Source:HGNC Symbol;Acc:HGNC:11142] | 3738 | 0.448 | 0.8184 | No |
| 38 | KLF6 | Kruppel like factor 6 [Source:HGNC Symbol;Acc:HGNC:2235] | 4799 | 0.264 | 0.7923 | No |
| 39 | TNFRSF11B | TNF receptor superfamily member 11b [Source:HGNC Symbol;Acc:HGNC:11909] | 5090 | 0.220 | 0.7866 | No |
| 40 | MCL1 | MCL1 apoptosis regulator, BCL2 family member [Source:HGNC Symbol;Acc:HGNC:6943] | 5624 | 0.142 | 0.7736 | No |
| 41 | DNAJB1 | DnaJ heat shock protein family (Hsp40) member B1 [Source:HGNC Symbol;Acc:HGNC:5270] | 5670 | 0.137 | 0.7737 | No |
| 42 | TIPARP | TCDD inducible poly(ADP-ribose) polymerase [Source:HGNC Symbol;Acc:HGNC:23696] | 5795 | 0.118 | 0.7715 | No |
| 43 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796] | 5920 | 0.104 | 0.7692 | No |
| 44 | CCN2 | cellular communication network factor 2 [Source:HGNC Symbol;Acc:HGNC:2500] | 6179 | 0.071 | 0.7629 | No |
| 45 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] | 7515 | 0.000 | 0.7269 | No |
| 46 | AREG | amphiregulin [Source:HGNC Symbol;Acc:HGNC:651] | 7611 | 0.000 | 0.7243 | No |
| 47 | DLX2 | distal-less homeobox 2 [Source:HGNC Symbol;Acc:HGNC:2915] | 16844 | 0.000 | 0.4750 | No |
| 48 | F2RL1 | F2R like trypsin receptor 1 [Source:HGNC Symbol;Acc:HGNC:3538] | 21188 | 0.000 | 0.3578 | No |
| 49 | EGR4 | early growth response 4 [Source:HGNC Symbol;Acc:HGNC:3241] | 22276 | 0.000 | 0.3284 | No |
| 50 | CCN1 | cellular communication network factor 1 [Source:HGNC Symbol;Acc:HGNC:2654] | 22967 | 0.000 | 0.3098 | No |
| 51 | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 [Source:HGNC Symbol;Acc:HGNC:7733] | 28773 | -0.152 | 0.1545 | No |
| 52 | GEM | GTP binding protein overexpressed in skeletal muscle [Source:HGNC Symbol;Acc:HGNC:4234] | 28985 | -0.176 | 0.1505 | No |
| 53 | NAP1L1 | nucleosome assembly protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:7637] | 29155 | -0.199 | 0.1479 | No |
| 54 | EPHA2 | EPH receptor A2 [Source:HGNC Symbol;Acc:HGNC:3386] | 30988 | -0.437 | 0.1027 | No |
| 55 | SPRED2 | sprouty related EVH1 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:17722] | 33763 | -1.052 | 0.0380 | No |
| 56 | BHLHE40 | basic helix-loop-helix family member e40 [Source:HGNC Symbol;Acc:HGNC:1046] | 35196 | -1.611 | 0.0150 | No |
| 57 | KLF10 | Kruppel like factor 10 [Source:HGNC Symbol;Acc:HGNC:11810] | 36962 | -3.715 | 0.0035 | No |