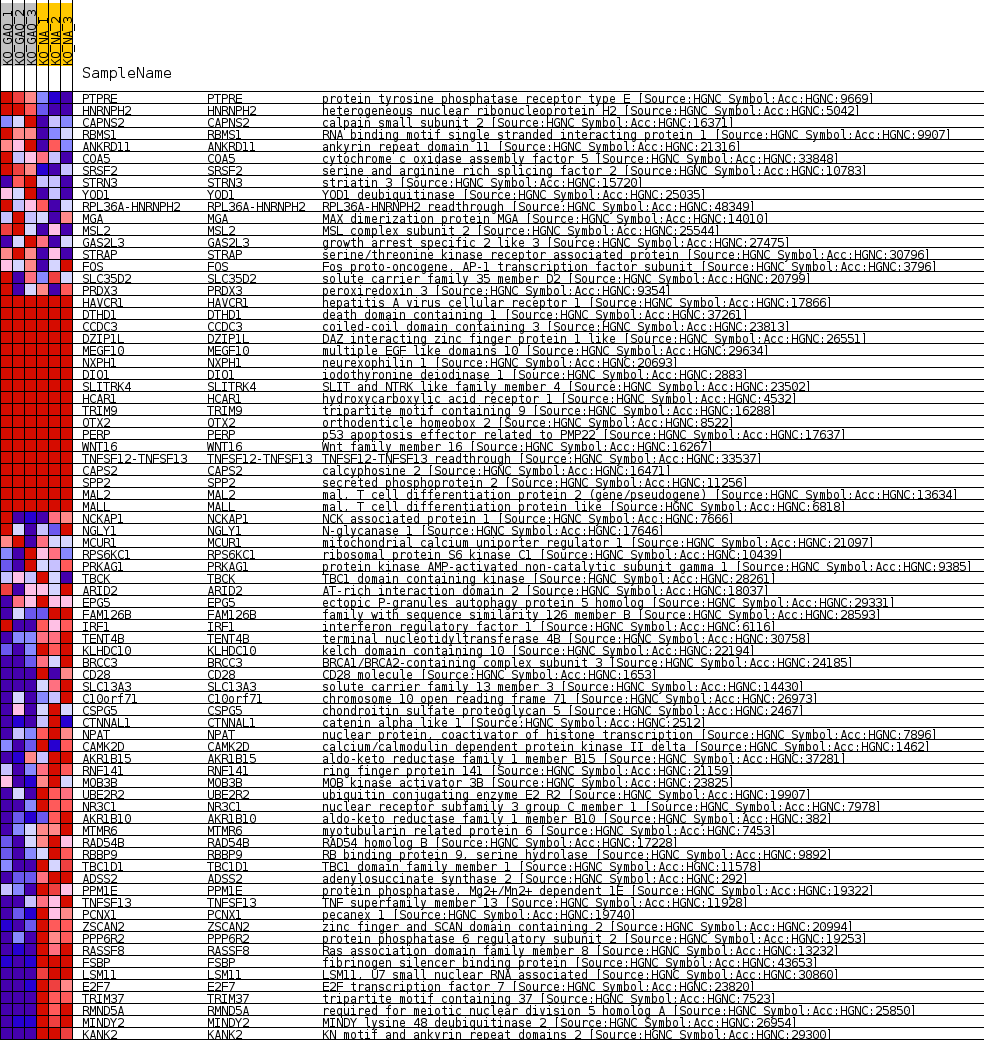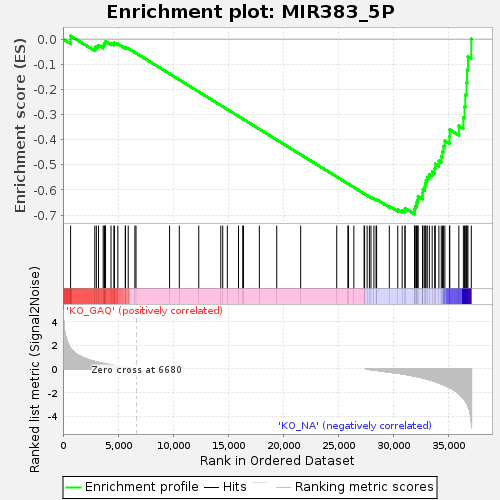
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR383_5P |
| Enrichment Score (ES) | -0.6958972 |
| Normalized Enrichment Score (NES) | -1.4393559 |
| Nominal p-value | 0.0048367595 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PTPRE | protein tyrosine phosphatase receptor type E [Source:HGNC Symbol;Acc:HGNC:9669] | 689 | 1.730 | 0.0126 | No |
| 2 | HNRNPH2 | heterogeneous nuclear ribonucleoprotein H2 [Source:HGNC Symbol;Acc:HGNC:5042] | 2886 | 0.620 | -0.0356 | No |
| 3 | CAPNS2 | calpain small subunit 2 [Source:HGNC Symbol;Acc:HGNC:16371] | 3027 | 0.584 | -0.0289 | No |
| 4 | RBMS1 | RNA binding motif single stranded interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:9907] | 3224 | 0.546 | -0.0243 | No |
| 5 | ANKRD11 | ankyrin repeat domain 11 [Source:HGNC Symbol;Acc:HGNC:21316] | 3647 | 0.464 | -0.0274 | No |
| 6 | COA5 | cytochrome c oxidase assembly factor 5 [Source:HGNC Symbol;Acc:HGNC:33848] | 3744 | 0.447 | -0.0219 | No |
| 7 | SRSF2 | serine and arginine rich splicing factor 2 [Source:HGNC Symbol;Acc:HGNC:10783] | 3782 | 0.441 | -0.0150 | No |
| 8 | STRN3 | striatin 3 [Source:HGNC Symbol;Acc:HGNC:15720] | 3862 | 0.426 | -0.0094 | No |
| 9 | YOD1 | YOD1 deubiquitinase [Source:HGNC Symbol;Acc:HGNC:25035] | 4359 | 0.336 | -0.0168 | No |
| 10 | RPL36A-HNRNPH2 | RPL36A-HNRNPH2 readthrough [Source:HGNC Symbol;Acc:HGNC:48349] | 4623 | 0.294 | -0.0186 | No |
| 11 | MGA | MAX dimerization protein MGA [Source:HGNC Symbol;Acc:HGNC:14010] | 4664 | 0.285 | -0.0145 | No |
| 12 | MSL2 | MSL complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:25544] | 4979 | 0.236 | -0.0188 | No |
| 13 | GAS2L3 | growth arrest specific 2 like 3 [Source:HGNC Symbol;Acc:HGNC:27475] | 5659 | 0.138 | -0.0346 | No |
| 14 | STRAP | serine/threonine kinase receptor associated protein [Source:HGNC Symbol;Acc:HGNC:30796] | 5663 | 0.138 | -0.0322 | No |
| 15 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796] | 5920 | 0.104 | -0.0373 | No |
| 16 | SLC35D2 | solute carrier family 35 member D2 [Source:HGNC Symbol;Acc:HGNC:20799] | 6525 | 0.024 | -0.0532 | No |
| 17 | PRDX3 | peroxiredoxin 3 [Source:HGNC Symbol;Acc:HGNC:9354] | 6627 | 0.008 | -0.0557 | No |
| 18 | HAVCR1 | hepatitis A virus cellular receptor 1 [Source:HGNC Symbol;Acc:HGNC:17866] | 9675 | 0.000 | -0.1381 | No |
| 19 | DTHD1 | death domain containing 1 [Source:HGNC Symbol;Acc:HGNC:37261] | 10558 | 0.000 | -0.1619 | No |
| 20 | CCDC3 | coiled-coil domain containing 3 [Source:HGNC Symbol;Acc:HGNC:23813] | 12324 | 0.000 | -0.2096 | No |
| 21 | DZIP1L | DAZ interacting zinc finger protein 1 like [Source:HGNC Symbol;Acc:HGNC:26551] | 14324 | 0.000 | -0.2636 | No |
| 22 | MEGF10 | multiple EGF like domains 10 [Source:HGNC Symbol;Acc:HGNC:29634] | 14501 | 0.000 | -0.2683 | No |
| 23 | NXPH1 | neurexophilin 1 [Source:HGNC Symbol;Acc:HGNC:20693] | 14918 | 0.000 | -0.2796 | No |
| 24 | DIO1 | iodothyronine deiodinase 1 [Source:HGNC Symbol;Acc:HGNC:2883] | 15933 | 0.000 | -0.3070 | No |
| 25 | SLITRK4 | SLIT and NTRK like family member 4 [Source:HGNC Symbol;Acc:HGNC:23502] | 16315 | 0.000 | -0.3173 | No |
| 26 | HCAR1 | hydroxycarboxylic acid receptor 1 [Source:HGNC Symbol;Acc:HGNC:4532] | 16387 | 0.000 | -0.3192 | No |
| 27 | TRIM9 | tripartite motif containing 9 [Source:HGNC Symbol;Acc:HGNC:16288] | 17825 | 0.000 | -0.3580 | No |
| 28 | OTX2 | orthodenticle homeobox 2 [Source:HGNC Symbol;Acc:HGNC:8522] | 19402 | 0.000 | -0.4006 | No |
| 29 | PERP | p53 apoptosis effector related to PMP22 [Source:HGNC Symbol;Acc:HGNC:17637] | 21576 | 0.000 | -0.4593 | No |
| 30 | WNT16 | Wnt family member 16 [Source:HGNC Symbol;Acc:HGNC:16267] | 24850 | 0.000 | -0.5477 | No |
| 31 | TNFSF12-TNFSF13 | TNFSF12-TNFSF13 readthrough [Source:HGNC Symbol;Acc:HGNC:33537] | 25860 | 0.000 | -0.5750 | No |
| 32 | CAPS2 | calcyphosine 2 [Source:HGNC Symbol;Acc:HGNC:16471] | 25923 | 0.000 | -0.5767 | No |
| 33 | SPP2 | secreted phosphoprotein 2 [Source:HGNC Symbol;Acc:HGNC:11256] | 26403 | 0.000 | -0.5896 | No |
| 34 | MAL2 | mal, T cell differentiation protein 2 (gene/pseudogene) [Source:HGNC Symbol;Acc:HGNC:13634] | 27332 | 0.000 | -0.6147 | No |
| 35 | MALL | mal, T cell differentiation protein like [Source:HGNC Symbol;Acc:HGNC:6818] | 27378 | 0.000 | -0.6159 | No |
| 36 | NCKAP1 | NCK associated protein 1 [Source:HGNC Symbol;Acc:HGNC:7666] | 27609 | -0.016 | -0.6218 | No |
| 37 | NGLY1 | N-glycanase 1 [Source:HGNC Symbol;Acc:HGNC:17646] | 27815 | -0.039 | -0.6267 | No |
| 38 | MCUR1 | mitochondrial calcium uniporter regulator 1 [Source:HGNC Symbol;Acc:HGNC:21097] | 27957 | -0.057 | -0.6295 | No |
| 39 | RPS6KC1 | ribosomal protein S6 kinase C1 [Source:HGNC Symbol;Acc:HGNC:10439] | 28225 | -0.087 | -0.6351 | No |
| 40 | PRKAG1 | protein kinase AMP-activated non-catalytic subunit gamma 1 [Source:HGNC Symbol;Acc:HGNC:9385] | 28421 | -0.110 | -0.6384 | No |
| 41 | TBCK | TBC1 domain containing kinase [Source:HGNC Symbol;Acc:HGNC:28261] | 28470 | -0.114 | -0.6376 | No |
| 42 | ARID2 | AT-rich interaction domain 2 [Source:HGNC Symbol;Acc:HGNC:18037] | 29621 | -0.256 | -0.6641 | No |
| 43 | EPG5 | ectopic P-granules autophagy protein 5 homolog [Source:HGNC Symbol;Acc:HGNC:29331] | 30389 | -0.353 | -0.6784 | No |
| 44 | FAM126B | family with sequence similarity 126 member B [Source:HGNC Symbol;Acc:HGNC:28593] | 30790 | -0.409 | -0.6819 | No |
| 45 | IRF1 | interferon regulatory factor 1 [Source:HGNC Symbol;Acc:HGNC:6116] | 31046 | -0.447 | -0.6807 | No |
| 46 | TENT4B | terminal nucleotidyltransferase 4B [Source:HGNC Symbol;Acc:HGNC:30758] | 31049 | -0.447 | -0.6727 | No |
| 47 | KLHDC10 | kelch domain containing 10 [Source:HGNC Symbol;Acc:HGNC:22194] | 31908 | -0.591 | -0.6853 | Yes |
| 48 | BRCC3 | BRCA1/BRCA2-containing complex subunit 3 [Source:HGNC Symbol;Acc:HGNC:24185] | 31926 | -0.594 | -0.6750 | Yes |
| 49 | CD28 | CD28 molecule [Source:HGNC Symbol;Acc:HGNC:1653] | 31973 | -0.605 | -0.6654 | Yes |
| 50 | SLC13A3 | solute carrier family 13 member 3 [Source:HGNC Symbol;Acc:HGNC:14430] | 32094 | -0.631 | -0.6572 | Yes |
| 51 | C10orf71 | chromosome 10 open reading frame 71 [Source:HGNC Symbol;Acc:HGNC:26973] | 32128 | -0.638 | -0.6466 | Yes |
| 52 | CSPG5 | chondroitin sulfate proteoglycan 5 [Source:HGNC Symbol;Acc:HGNC:2467] | 32212 | -0.657 | -0.6370 | Yes |
| 53 | CTNNAL1 | catenin alpha like 1 [Source:HGNC Symbol;Acc:HGNC:2512] | 32230 | -0.660 | -0.6256 | Yes |
| 54 | NPAT | nuclear protein, coactivator of histone transcription [Source:HGNC Symbol;Acc:HGNC:7896] | 32636 | -0.743 | -0.6232 | Yes |
| 55 | CAMK2D | calcium/calmodulin dependent protein kinase II delta [Source:HGNC Symbol;Acc:HGNC:1462] | 32651 | -0.746 | -0.6101 | Yes |
| 56 | AKR1B15 | aldo-keto reductase family 1 member B15 [Source:HGNC Symbol;Acc:HGNC:37281] | 32667 | -0.750 | -0.5970 | Yes |
| 57 | RNF141 | ring finger protein 141 [Source:HGNC Symbol;Acc:HGNC:21159] | 32842 | -0.795 | -0.5874 | Yes |
| 58 | MOB3B | MOB kinase activator 3B [Source:HGNC Symbol;Acc:HGNC:23825] | 32877 | -0.803 | -0.5738 | Yes |
| 59 | UBE2R2 | ubiquitin conjugating enzyme E2 R2 [Source:HGNC Symbol;Acc:HGNC:19907] | 32952 | -0.819 | -0.5611 | Yes |
| 60 | NR3C1 | nuclear receptor subfamily 3 group C member 1 [Source:HGNC Symbol;Acc:HGNC:7978] | 33067 | -0.850 | -0.5488 | Yes |
| 61 | AKR1B10 | aldo-keto reductase family 1 member B10 [Source:HGNC Symbol;Acc:HGNC:382] | 33258 | -0.904 | -0.5377 | Yes |
| 62 | MTMR6 | myotubularin related protein 6 [Source:HGNC Symbol;Acc:HGNC:7453] | 33514 | -0.972 | -0.5271 | Yes |
| 63 | RAD54B | RAD54 homolog B [Source:HGNC Symbol;Acc:HGNC:17228] | 33743 | -1.046 | -0.5144 | Yes |
| 64 | RBBP9 | RB binding protein 9, serine hydrolase [Source:HGNC Symbol;Acc:HGNC:9892] | 33794 | -1.061 | -0.4966 | Yes |
| 65 | TBC1D1 | TBC1 domain family member 1 [Source:HGNC Symbol;Acc:HGNC:11578] | 34121 | -1.172 | -0.4843 | Yes |
| 66 | ADSS2 | adenylosuccinate synthase 2 [Source:HGNC Symbol;Acc:HGNC:292] | 34338 | -1.246 | -0.4677 | Yes |
| 67 | PPM1E | protein phosphatase, Mg2+/Mn2+ dependent 1E [Source:HGNC Symbol;Acc:HGNC:19322] | 34445 | -1.290 | -0.4473 | Yes |
| 68 | TNFSF13 | TNF superfamily member 13 [Source:HGNC Symbol;Acc:HGNC:11928] | 34534 | -1.326 | -0.4258 | Yes |
| 69 | PCNX1 | pecanex 1 [Source:HGNC Symbol;Acc:HGNC:19740] | 34666 | -1.377 | -0.4045 | Yes |
| 70 | ZSCAN2 | zinc finger and SCAN domain containing 2 [Source:HGNC Symbol;Acc:HGNC:20994] | 35080 | -1.551 | -0.3877 | Yes |
| 71 | PPP6R2 | protein phosphatase 6 regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:19253] | 35110 | -1.570 | -0.3602 | Yes |
| 72 | RASSF8 | Ras association domain family member 8 [Source:HGNC Symbol;Acc:HGNC:13232] | 35935 | -2.078 | -0.3451 | Yes |
| 73 | FSBP | fibrinogen silencer binding protein [Source:HGNC Symbol;Acc:HGNC:43653] | 36344 | -2.456 | -0.3119 | Yes |
| 74 | LSM11 | LSM11, U7 small nuclear RNA associated [Source:HGNC Symbol;Acc:HGNC:30860] | 36453 | -2.577 | -0.2683 | Yes |
| 75 | E2F7 | E2F transcription factor 7 [Source:HGNC Symbol;Acc:HGNC:23820] | 36519 | -2.672 | -0.2220 | Yes |
| 76 | TRIM37 | tripartite motif containing 37 [Source:HGNC Symbol;Acc:HGNC:7523] | 36633 | -2.822 | -0.1742 | Yes |
| 77 | RMND5A | required for meiotic nuclear division 5 homolog A [Source:HGNC Symbol;Acc:HGNC:25850] | 36681 | -2.900 | -0.1232 | Yes |
| 78 | MINDY2 | MINDY lysine 48 deubiquitinase 2 [Source:HGNC Symbol;Acc:HGNC:26954] | 36760 | -3.085 | -0.0697 | Yes |
| 79 | KANK2 | KN motif and ankyrin repeat domains 2 [Source:HGNC Symbol;Acc:HGNC:29300] | 37065 | -4.363 | 0.0007 | Yes |