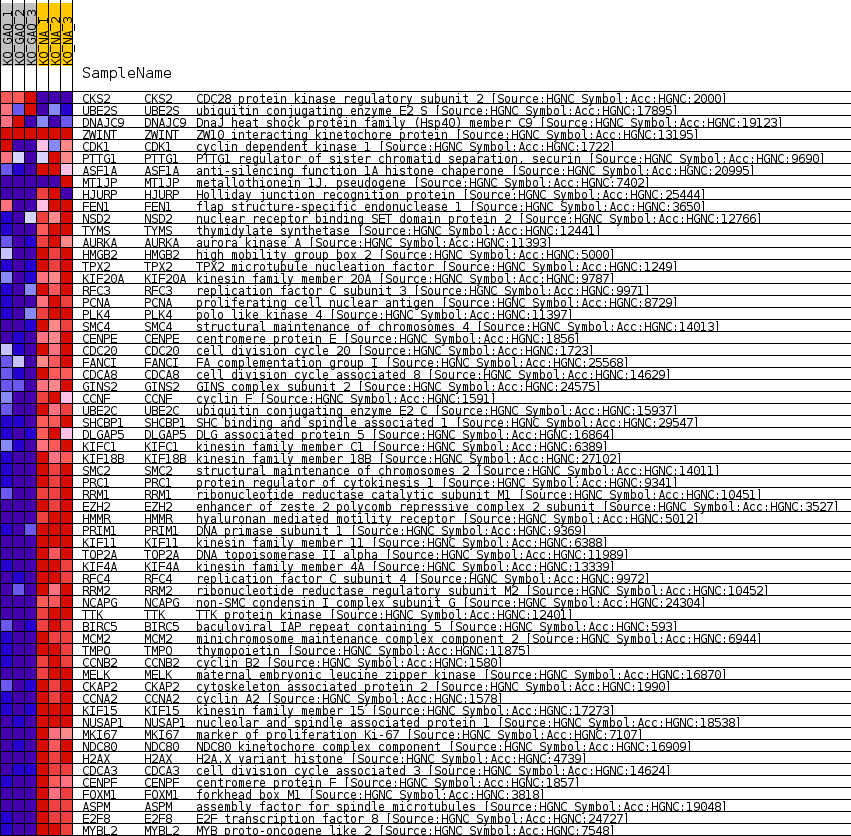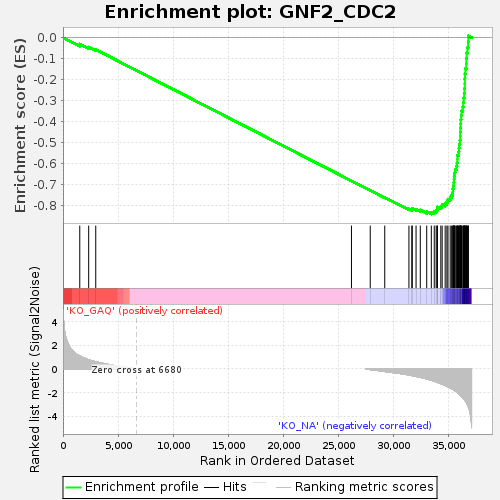
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_CDC2 |
| Enrichment Score (ES) | -0.8427867 |
| Normalized Enrichment Score (NES) | -1.7139286 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1522 | 1.100 | -0.0310 | No |
| 2 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 2332 | 0.774 | -0.0457 | No |
| 3 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 2973 | 0.593 | -0.0575 | No |
| 4 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26185 | 0.000 | -0.6843 | No |
| 5 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27890 | -0.049 | -0.7299 | No |
| 6 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 29208 | -0.205 | -0.7636 | No |
| 7 | ASF1A | anti-silencing function 1A histone chaperone [Source:HGNC Symbol;Acc:HGNC:20995] | 31404 | -0.506 | -0.8182 | No |
| 8 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 31668 | -0.553 | -0.8202 | No |
| 9 | HJURP | Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] | 31702 | -0.560 | -0.8159 | No |
| 10 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 32050 | -0.618 | -0.8196 | No |
| 11 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 32434 | -0.702 | -0.8235 | No |
| 12 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 33018 | -0.836 | -0.8316 | No |
| 13 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 33435 | -0.948 | -0.8341 | Yes |
| 14 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33703 | -1.035 | -0.8317 | Yes |
| 15 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 33875 | -1.087 | -0.8263 | Yes |
| 16 | KIF20A | kinesin family member 20A [Source:HGNC Symbol;Acc:HGNC:9787] | 33965 | -1.117 | -0.8185 | Yes |
| 17 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33988 | -1.124 | -0.8087 | Yes |
| 18 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 34299 | -1.231 | -0.8057 | Yes |
| 19 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34419 | -1.276 | -0.7972 | Yes |
| 20 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34692 | -1.386 | -0.7918 | Yes |
| 21 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 34850 | -1.453 | -0.7826 | Yes |
| 22 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34948 | -1.490 | -0.7715 | Yes |
| 23 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35150 | -1.587 | -0.7623 | Yes |
| 24 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 35283 | -1.667 | -0.7505 | Yes |
| 25 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35385 | -1.715 | -0.7375 | Yes |
| 26 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 35395 | -1.722 | -0.7218 | Yes |
| 27 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35447 | -1.747 | -0.7071 | Yes |
| 28 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 35471 | -1.761 | -0.6915 | Yes |
| 29 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35498 | -1.774 | -0.6759 | Yes |
| 30 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 35510 | -1.778 | -0.6598 | Yes |
| 31 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 35559 | -1.810 | -0.6444 | Yes |
| 32 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35602 | -1.838 | -0.6286 | Yes |
| 33 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 35709 | -1.910 | -0.6139 | Yes |
| 34 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35769 | -1.949 | -0.5975 | Yes |
| 35 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 35806 | -1.978 | -0.5803 | Yes |
| 36 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35808 | -1.979 | -0.5621 | Yes |
| 37 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35905 | -2.049 | -0.5458 | Yes |
| 38 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35949 | -2.088 | -0.5277 | Yes |
| 39 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35971 | -2.105 | -0.5089 | Yes |
| 40 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 36027 | -2.160 | -0.4905 | Yes |
| 41 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36061 | -2.182 | -0.4713 | Yes |
| 42 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36065 | -2.187 | -0.4512 | Yes |
| 43 | NCAPG | non-SMC condensin I complex subunit G [Source:HGNC Symbol;Acc:HGNC:24304] | 36078 | -2.201 | -0.4313 | Yes |
| 44 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36102 | -2.226 | -0.4114 | Yes |
| 45 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36111 | -2.235 | -0.3910 | Yes |
| 46 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36147 | -2.266 | -0.3711 | Yes |
| 47 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36174 | -2.293 | -0.3507 | Yes |
| 48 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36291 | -2.397 | -0.3317 | Yes |
| 49 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36336 | -2.447 | -0.3104 | Yes |
| 50 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 36363 | -2.471 | -0.2883 | Yes |
| 51 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36418 | -2.536 | -0.2664 | Yes |
| 52 | KIF15 | kinesin family member 15 [Source:HGNC Symbol;Acc:HGNC:17273] | 36451 | -2.572 | -0.2436 | Yes |
| 53 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36471 | -2.598 | -0.2202 | Yes |
| 54 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36474 | -2.605 | -0.1962 | Yes |
| 55 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36492 | -2.640 | -0.1724 | Yes |
| 56 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36530 | -2.698 | -0.1485 | Yes |
| 57 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36614 | -2.799 | -0.1250 | Yes |
| 58 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36617 | -2.804 | -0.0992 | Yes |
| 59 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36641 | -2.835 | -0.0737 | Yes |
| 60 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36716 | -2.984 | -0.0482 | Yes |
| 61 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36779 | -3.125 | -0.0211 | Yes |
| 62 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36799 | -3.202 | 0.0079 | Yes |