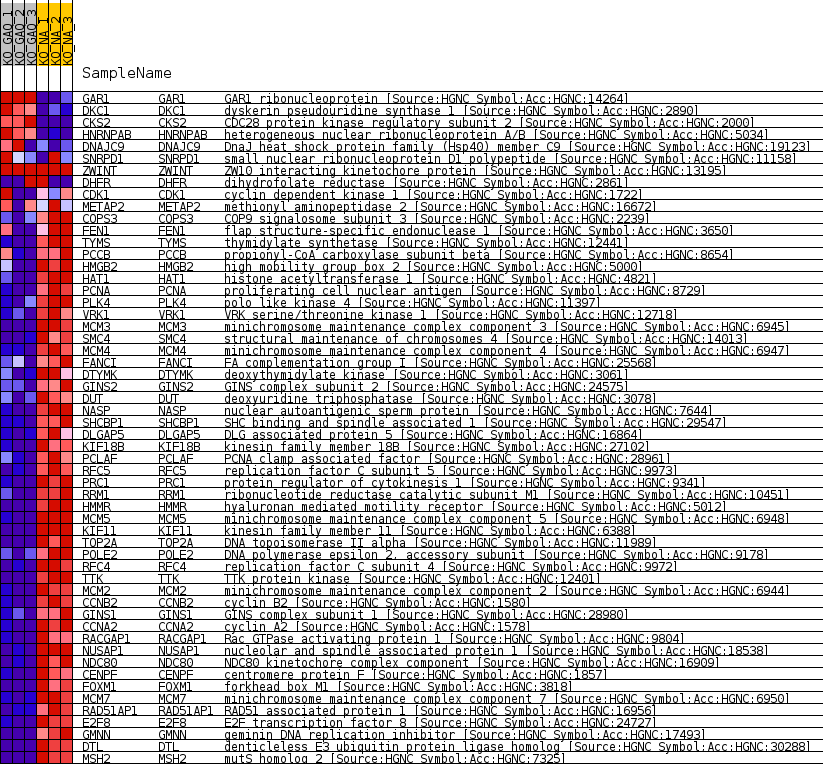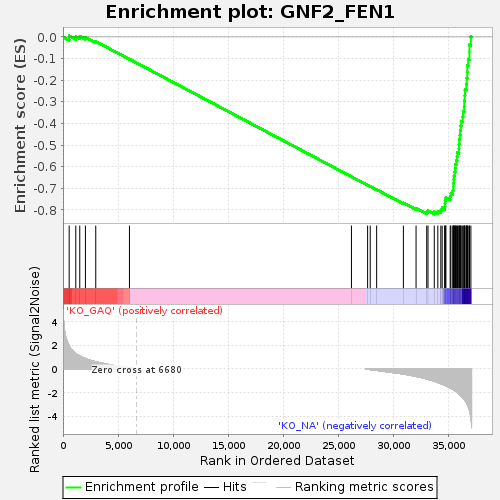
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_FEN1 |
| Enrichment Score (ES) | -0.819662 |
| Normalized Enrichment Score (NES) | -1.6233618 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0016696672 |
| FWER p-Value | 0.036 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GAR1 | GAR1 ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:14264] | 560 | 1.953 | 0.0048 | No |
| 2 | DKC1 | dyskerin pseudouridine synthase 1 [Source:HGNC Symbol;Acc:HGNC:2890] | 1160 | 1.291 | 0.0017 | No |
| 3 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1522 | 1.100 | 0.0032 | No |
| 4 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B [Source:HGNC Symbol;Acc:HGNC:5034] | 2035 | 0.878 | -0.0017 | No |
| 5 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 2973 | 0.593 | -0.0209 | No |
| 6 | SNRPD1 | small nuclear ribonucleoprotein D1 polypeptide [Source:HGNC Symbol;Acc:HGNC:11158] | 6032 | 0.088 | -0.1026 | No |
| 7 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26185 | 0.000 | -0.6468 | No |
| 8 | DHFR | dihydrofolate reductase [Source:HGNC Symbol;Acc:HGNC:2861] | 27652 | -0.021 | -0.6861 | No |
| 9 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27890 | -0.049 | -0.6920 | No |
| 10 | METAP2 | methionyl aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:16672] | 28473 | -0.115 | -0.7066 | No |
| 11 | COPS3 | COP9 signalosome subunit 3 [Source:HGNC Symbol;Acc:HGNC:2239] | 30900 | -0.427 | -0.7677 | No |
| 12 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 32050 | -0.618 | -0.7925 | No |
| 13 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 33018 | -0.836 | -0.8101 | Yes |
| 14 | PCCB | propionyl-CoA carboxylase subunit beta [Source:HGNC Symbol;Acc:HGNC:8654] | 33134 | -0.868 | -0.8043 | Yes |
| 15 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33703 | -1.035 | -0.8091 | Yes |
| 16 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 34026 | -1.138 | -0.8062 | Yes |
| 17 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 34299 | -1.231 | -0.8010 | Yes |
| 18 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34419 | -1.276 | -0.7913 | Yes |
| 19 | VRK1 | VRK serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:12718] | 34657 | -1.375 | -0.7836 | Yes |
| 20 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34678 | -1.382 | -0.7701 | Yes |
| 21 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34692 | -1.386 | -0.7563 | Yes |
| 22 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34760 | -1.422 | -0.7437 | Yes |
| 23 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35150 | -1.587 | -0.7380 | Yes |
| 24 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 35211 | -1.621 | -0.7231 | Yes |
| 25 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35385 | -1.715 | -0.7103 | Yes |
| 26 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 35438 | -1.743 | -0.6940 | Yes |
| 27 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35440 | -1.743 | -0.6762 | Yes |
| 28 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 35471 | -1.761 | -0.6591 | Yes |
| 29 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35498 | -1.774 | -0.6418 | Yes |
| 30 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 35559 | -1.810 | -0.6249 | Yes |
| 31 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35608 | -1.839 | -0.6075 | Yes |
| 32 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 35628 | -1.853 | -0.5891 | Yes |
| 33 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 35709 | -1.910 | -0.5718 | Yes |
| 34 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35769 | -1.949 | -0.5536 | Yes |
| 35 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35808 | -1.979 | -0.5345 | Yes |
| 36 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35939 | -2.080 | -0.5168 | Yes |
| 37 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35949 | -2.088 | -0.4958 | Yes |
| 38 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35971 | -2.105 | -0.4749 | Yes |
| 39 | POLE2 | DNA polymerase epsilon 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9178] | 36028 | -2.161 | -0.4544 | Yes |
| 40 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36061 | -2.182 | -0.4330 | Yes |
| 41 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36102 | -2.226 | -0.4114 | Yes |
| 42 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36147 | -2.266 | -0.3895 | Yes |
| 43 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36291 | -2.397 | -0.3690 | Yes |
| 44 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36323 | -2.430 | -0.3451 | Yes |
| 45 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36418 | -2.536 | -0.3218 | Yes |
| 46 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36429 | -2.546 | -0.2961 | Yes |
| 47 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36471 | -2.598 | -0.2708 | Yes |
| 48 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36492 | -2.640 | -0.2444 | Yes |
| 49 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36617 | -2.804 | -0.2192 | Yes |
| 50 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36641 | -2.835 | -0.1909 | Yes |
| 51 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36694 | -2.936 | -0.1624 | Yes |
| 52 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 36707 | -2.964 | -0.1326 | Yes |
| 53 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36779 | -3.125 | -0.1027 | Yes |
| 54 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36895 | -3.442 | -0.0707 | Yes |
| 55 | DTL | denticleless E3 ubiquitin protein ligase homolog [Source:HGNC Symbol;Acc:HGNC:30288] | 36896 | -3.444 | -0.0356 | Yes |
| 56 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 37028 | -4.011 | 0.0017 | Yes |