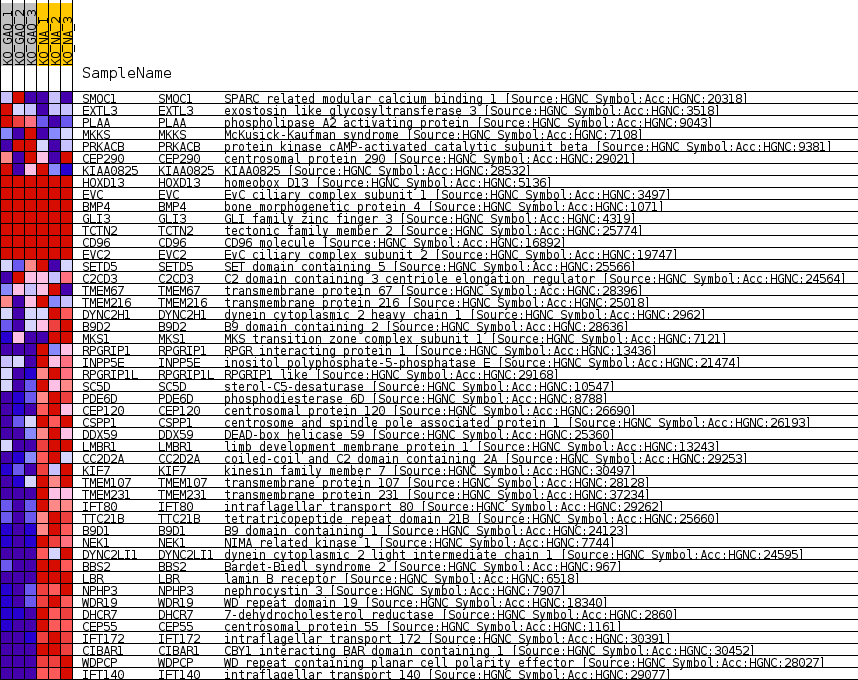
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HP_POSTAXIAL_FOOT_POLYDACTYLY |
| Enrichment Score (ES) | -0.8049504 |
| Normalized Enrichment Score (NES) | -1.5946563 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07457996 |
| FWER p-Value | 0.758 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SMOC1 | SPARC related modular calcium binding 1 [Source:HGNC Symbol;Acc:HGNC:20318] | 3380 | 0.512 | -0.0820 | No |
| 2 | EXTL3 | exostosin like glycosyltransferase 3 [Source:HGNC Symbol;Acc:HGNC:3518] | 3757 | 0.445 | -0.0841 | No |
| 3 | PLAA | phospholipase A2 activating protein [Source:HGNC Symbol;Acc:HGNC:9043] | 4034 | 0.390 | -0.0845 | No |
| 4 | MKKS | McKusick-Kaufman syndrome [Source:HGNC Symbol;Acc:HGNC:7108] | 4079 | 0.383 | -0.0787 | No |
| 5 | PRKACB | protein kinase cAMP-activated catalytic subunit beta [Source:HGNC Symbol;Acc:HGNC:9381] | 4092 | 0.381 | -0.0722 | No |
| 6 | CEP290 | centrosomal protein 290 [Source:HGNC Symbol;Acc:HGNC:29021] | 6423 | 0.040 | -0.1344 | No |
| 7 | KIAA0825 | KIAA0825 [Source:HGNC Symbol;Acc:HGNC:28532] | 6673 | 0.000 | -0.1411 | No |
| 8 | HOXD13 | homeobox D13 [Source:HGNC Symbol;Acc:HGNC:5136] | 13406 | 0.000 | -0.3228 | No |
| 9 | EVC | EvC ciliary complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:3497] | 16598 | 0.000 | -0.4090 | No |
| 10 | BMP4 | bone morphogenetic protein 4 [Source:HGNC Symbol;Acc:HGNC:1071] | 19060 | 0.000 | -0.4754 | No |
| 11 | GLI3 | GLI family zinc finger 3 [Source:HGNC Symbol;Acc:HGNC:4319] | 21772 | 0.000 | -0.5486 | No |
| 12 | TCTN2 | tectonic family member 2 [Source:HGNC Symbol;Acc:HGNC:25774] | 22453 | 0.000 | -0.5669 | No |
| 13 | CD96 | CD96 molecule [Source:HGNC Symbol;Acc:HGNC:16892] | 23113 | 0.000 | -0.5847 | No |
| 14 | EVC2 | EvC ciliary complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:19747] | 23491 | 0.000 | -0.5949 | No |
| 15 | SETD5 | SET domain containing 5 [Source:HGNC Symbol;Acc:HGNC:25566] | 27836 | -0.041 | -0.7114 | No |
| 16 | C2CD3 | C2 domain containing 3 centriole elongation regulator [Source:HGNC Symbol;Acc:HGNC:24564] | 27931 | -0.055 | -0.7130 | No |
| 17 | TMEM67 | transmembrane protein 67 [Source:HGNC Symbol;Acc:HGNC:28396] | 28693 | -0.142 | -0.7310 | No |
| 18 | TMEM216 | transmembrane protein 216 [Source:HGNC Symbol;Acc:HGNC:25018] | 29417 | -0.231 | -0.7463 | No |
| 19 | DYNC2H1 | dynein cytoplasmic 2 heavy chain 1 [Source:HGNC Symbol;Acc:HGNC:2962] | 30395 | -0.354 | -0.7663 | No |
| 20 | B9D2 | B9 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28636] | 31315 | -0.487 | -0.7823 | No |
| 21 | MKS1 | MKS transition zone complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:7121] | 31627 | -0.544 | -0.7808 | No |
| 22 | RPGRIP1 | RPGR interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:13436] | 32522 | -0.717 | -0.7920 | Yes |
| 23 | INPP5E | inositol polyphosphate-5-phosphatase E [Source:HGNC Symbol;Acc:HGNC:21474] | 32609 | -0.737 | -0.7810 | Yes |
| 24 | RPGRIP1L | RPGRIP1 like [Source:HGNC Symbol;Acc:HGNC:29168] | 32917 | -0.813 | -0.7745 | Yes |
| 25 | SC5D | sterol-C5-desaturase [Source:HGNC Symbol;Acc:HGNC:10547] | 33659 | -1.018 | -0.7761 | Yes |
| 26 | PDE6D | phosphodiesterase 6D [Source:HGNC Symbol;Acc:HGNC:8788] | 33824 | -1.071 | -0.7612 | Yes |
| 27 | CEP120 | centrosomal protein 120 [Source:HGNC Symbol;Acc:HGNC:26690] | 34289 | -1.229 | -0.7515 | Yes |
| 28 | CSPP1 | centrosome and spindle pole associated protein 1 [Source:HGNC Symbol;Acc:HGNC:26193] | 34470 | -1.301 | -0.7328 | Yes |
| 29 | DDX59 | DEAD-box helicase 59 [Source:HGNC Symbol;Acc:HGNC:25360] | 34579 | -1.345 | -0.7114 | Yes |
| 30 | LMBR1 | limb development membrane protein 1 [Source:HGNC Symbol;Acc:HGNC:13243] | 34650 | -1.373 | -0.6884 | Yes |
| 31 | CC2D2A | coiled-coil and C2 domain containing 2A [Source:HGNC Symbol;Acc:HGNC:29253] | 34797 | -1.435 | -0.6664 | Yes |
| 32 | KIF7 | kinesin family member 7 [Source:HGNC Symbol;Acc:HGNC:30497] | 34814 | -1.441 | -0.6407 | Yes |
| 33 | TMEM107 | transmembrane protein 107 [Source:HGNC Symbol;Acc:HGNC:28128] | 34979 | -1.506 | -0.6179 | Yes |
| 34 | TMEM231 | transmembrane protein 231 [Source:HGNC Symbol;Acc:HGNC:37234] | 35114 | -1.572 | -0.5931 | Yes |
| 35 | IFT80 | intraflagellar transport 80 [Source:HGNC Symbol;Acc:HGNC:29262] | 35248 | -1.645 | -0.5669 | Yes |
| 36 | TTC21B | tetratricopeptide repeat domain 21B [Source:HGNC Symbol;Acc:HGNC:25660] | 35417 | -1.733 | -0.5401 | Yes |
| 37 | B9D1 | B9 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:24123] | 35439 | -1.743 | -0.5091 | Yes |
| 38 | NEK1 | NIMA related kinase 1 [Source:HGNC Symbol;Acc:HGNC:7744] | 35607 | -1.839 | -0.4804 | Yes |
| 39 | DYNC2LI1 | dynein cytoplasmic 2 light intermediate chain 1 [Source:HGNC Symbol;Acc:HGNC:24595] | 35638 | -1.866 | -0.4474 | Yes |
| 40 | BBS2 | Bardet-Biedl syndrome 2 [Source:HGNC Symbol;Acc:HGNC:967] | 35829 | -1.996 | -0.4164 | Yes |
| 41 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 35988 | -2.120 | -0.3823 | Yes |
| 42 | NPHP3 | nephrocystin 3 [Source:HGNC Symbol;Acc:HGNC:7907] | 36434 | -2.551 | -0.3482 | Yes |
| 43 | WDR19 | WD repeat domain 19 [Source:HGNC Symbol;Acc:HGNC:18340] | 36496 | -2.646 | -0.3020 | Yes |
| 44 | DHCR7 | 7-dehydrocholesterol reductase [Source:HGNC Symbol;Acc:HGNC:2860] | 36509 | -2.655 | -0.2542 | Yes |
| 45 | CEP55 | centrosomal protein 55 [Source:HGNC Symbol;Acc:HGNC:1161] | 36520 | -2.674 | -0.2061 | Yes |
| 46 | IFT172 | intraflagellar transport 172 [Source:HGNC Symbol;Acc:HGNC:30391] | 36540 | -2.715 | -0.1575 | Yes |
| 47 | CIBAR1 | CBY1 interacting BAR domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30452] | 36603 | -2.781 | -0.1088 | Yes |
| 48 | WDPCP | WD repeat containing planar cell polarity effector [Source:HGNC Symbol;Acc:HGNC:28027] | 36813 | -3.234 | -0.0560 | Yes |
| 49 | IFT140 | intraflagellar transport 140 [Source:HGNC Symbol;Acc:HGNC:29077] | 36915 | -3.505 | 0.0047 | Yes |