Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_GAQ_versus_KO_NA.greenbergcls.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | RB_P107_DN.V1_DN |
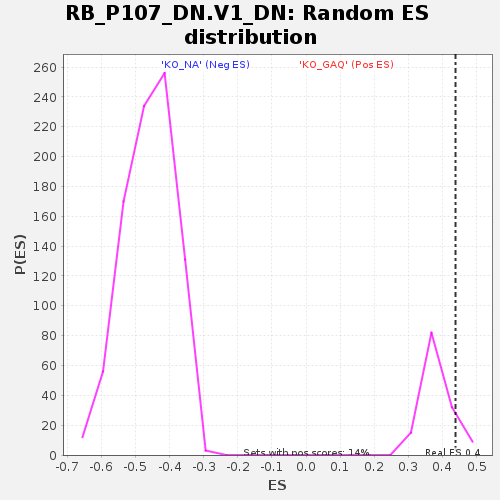
| Enrichment Score (ES) | 0.43826157 |
| Normalized Enrichment Score (NES) | 1.143134 |
| Nominal p-value | 0.10144927 |
| FDR q-value | 0.25504306 |
| FWER p-Value | 0.907 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PLK3 | polo like kinase 3 [Source:HGNC Symbol;Acc:HGNC:2154] | 33 | 4.365 | 0.0375 | Yes |
| 2 | EDN1 | endothelin 1 [Source:HGNC Symbol;Acc:HGNC:3176] | 44 | 4.198 | 0.0742 | Yes |
| 3 | MYO1D | myosin ID [Source:HGNC Symbol;Acc:HGNC:7598] | 88 | 3.623 | 0.1049 | Yes |
| 4 | GLRX | glutaredoxin [Source:HGNC Symbol;Acc:HGNC:4330] | 138 | 3.244 | 0.1321 | Yes |
| 5 | POR | cytochrome p450 oxidoreductase [Source:HGNC Symbol;Acc:HGNC:9208] | 171 | 3.055 | 0.1581 | Yes |
| 6 | BLCAP | BLCAP apoptosis inducing factor [Source:HGNC Symbol;Acc:HGNC:1055] | 215 | 2.838 | 0.1819 | Yes |
| 7 | GNG12 | G protein subunit gamma 12 [Source:HGNC Symbol;Acc:HGNC:19663] | 437 | 2.175 | 0.1950 | Yes |
| 8 | TMA16 | translation machinery associated 16 homolog [Source:HGNC Symbol;Acc:HGNC:25638] | 586 | 1.906 | 0.2078 | Yes |
| 9 | RARS1 | arginyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:9870] | 599 | 1.878 | 0.2240 | Yes |
| 10 | CHAC1 | ChaC glutathione specific gamma-glutamylcyclotransferase 1 [Source:HGNC Symbol;Acc:HGNC:28680] | 682 | 1.742 | 0.2371 | Yes |
| 11 | PTPRE | protein tyrosine phosphatase receptor type E [Source:HGNC Symbol;Acc:HGNC:9669] | 689 | 1.730 | 0.2522 | Yes |
| 12 | LPGAT1 | lysophosphatidylglycerol acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:28985] | 801 | 1.597 | 0.2632 | Yes |
| 13 | ANXA1 | annexin A1 [Source:HGNC Symbol;Acc:HGNC:533] | 913 | 1.493 | 0.2733 | Yes |
| 14 | NARS1 | asparaginyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:7643] | 944 | 1.459 | 0.2854 | Yes |
| 15 | TLR2 | toll like receptor 2 [Source:HGNC Symbol;Acc:HGNC:11848] | 1022 | 1.395 | 0.2956 | Yes |
| 16 | PMM2 | phosphomannomutase 2 [Source:HGNC Symbol;Acc:HGNC:9115] | 1053 | 1.372 | 0.3068 | Yes |
| 17 | DAD1 | defender against cell death 1 [Source:HGNC Symbol;Acc:HGNC:2664] | 1312 | 1.192 | 0.3103 | Yes |
| 18 | HSPH1 | heat shock protein family H (Hsp110) member 1 [Source:HGNC Symbol;Acc:HGNC:16969] | 1383 | 1.165 | 0.3187 | Yes |
| 19 | TDRD7 | tudor domain containing 7 [Source:HGNC Symbol;Acc:HGNC:30831] | 1445 | 1.135 | 0.3270 | Yes |
| 20 | SLIRP | SRA stem-loop interacting RNA binding protein [Source:HGNC Symbol;Acc:HGNC:20495] | 1467 | 1.127 | 0.3363 | Yes |
| 21 | DYNLL1 | dynein light chain LC8-type 1 [Source:HGNC Symbol;Acc:HGNC:15476] | 1677 | 1.024 | 0.3397 | Yes |
| 22 | POMP | proteasome maturation protein [Source:HGNC Symbol;Acc:HGNC:20330] | 1693 | 1.014 | 0.3482 | Yes |
| 23 | ELOVL1 | ELOVL fatty acid elongase 1 [Source:HGNC Symbol;Acc:HGNC:14418] | 1741 | 0.987 | 0.3556 | Yes |
| 24 | NFE2L1 | nuclear factor, erythroid 2 like 1 [Source:HGNC Symbol;Acc:HGNC:7781] | 2150 | 0.840 | 0.3520 | Yes |
| 25 | BCCIP | BRCA2 and CDKN1A interacting protein [Source:HGNC Symbol;Acc:HGNC:978] | 2245 | 0.801 | 0.3565 | Yes |
| 26 | TXN2 | thioredoxin 2 [Source:HGNC Symbol;Acc:HGNC:17772] | 2253 | 0.800 | 0.3633 | Yes |
| 27 | TARS1 | threonyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:11572] | 2275 | 0.792 | 0.3697 | Yes |
| 28 | APTX | aprataxin [Source:HGNC Symbol;Acc:HGNC:15984] | 2336 | 0.773 | 0.3749 | Yes |
| 29 | BSDC1 | BSD domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25501] | 2362 | 0.766 | 0.3810 | Yes |
| 30 | UBE2G2 | ubiquitin conjugating enzyme E2 G2 [Source:HGNC Symbol;Acc:HGNC:12483] | 2638 | 0.680 | 0.3795 | Yes |
| 31 | PGD | phosphogluconate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:8891] | 2710 | 0.663 | 0.3834 | Yes |
| 32 | UBA5 | ubiquitin like modifier activating enzyme 5 [Source:HGNC Symbol;Acc:HGNC:23230] | 2741 | 0.655 | 0.3884 | Yes |
| 33 | RBM8A | RNA binding motif protein 8A [Source:HGNC Symbol;Acc:HGNC:9905] | 2859 | 0.628 | 0.3907 | Yes |
| 34 | NFE2L2 | nuclear factor, erythroid 2 like 2 [Source:HGNC Symbol;Acc:HGNC:7782] | 2907 | 0.613 | 0.3948 | Yes |
| 35 | CRABP2 | cellular retinoic acid binding protein 2 [Source:HGNC Symbol;Acc:HGNC:2339] | 2971 | 0.594 | 0.3984 | Yes |
| 36 | PINX1 | PIN2 (TERF1) interacting telomerase inhibitor 1 [Source:HGNC Symbol;Acc:HGNC:30046] | 3048 | 0.579 | 0.4014 | Yes |
| 37 | AARS1 | alanyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:20] | 3074 | 0.576 | 0.4058 | Yes |
| 38 | EFL1 | elongation factor like GTPase 1 [Source:HGNC Symbol;Acc:HGNC:25789] | 3153 | 0.561 | 0.4086 | Yes |
| 39 | NRBP1 | nuclear receptor binding protein 1 [Source:HGNC Symbol;Acc:HGNC:7993] | 3156 | 0.560 | 0.4135 | Yes |
| 40 | HSD17B12 | hydroxysteroid 17-beta dehydrogenase 12 [Source:HGNC Symbol;Acc:HGNC:18646] | 3261 | 0.537 | 0.4154 | Yes |
| 41 | UNC119B | unc-119 lipid binding chaperone B [Source:HGNC Symbol;Acc:HGNC:16488] | 3288 | 0.532 | 0.4194 | Yes |
| 42 | RNF25 | ring finger protein 25 [Source:HGNC Symbol;Acc:HGNC:14662] | 3293 | 0.531 | 0.4239 | Yes |
| 43 | CAP1 | cyclase associated actin cytoskeleton regulatory protein 1 [Source:HGNC Symbol;Acc:HGNC:20040] | 3305 | 0.528 | 0.4283 | Yes |
| 44 | UROS | uroporphyrinogen III synthase [Source:HGNC Symbol;Acc:HGNC:12592] | 3491 | 0.491 | 0.4276 | Yes |
| 45 | ARPC1B | actin related protein 2/3 complex subunit 1B [Source:HGNC Symbol;Acc:HGNC:704] | 3674 | 0.459 | 0.4267 | Yes |
| 46 | COA5 | cytochrome c oxidase assembly factor 5 [Source:HGNC Symbol;Acc:HGNC:33848] | 3744 | 0.447 | 0.4288 | Yes |
| 47 | CLIP4 | CAP-Gly domain containing linker protein family member 4 [Source:HGNC Symbol;Acc:HGNC:26108] | 3747 | 0.447 | 0.4326 | Yes |
| 48 | DENND11 | DENN domain containing 11 [Source:HGNC Symbol;Acc:HGNC:29472] | 3823 | 0.434 | 0.4344 | Yes |
| 49 | ARL1 | ADP ribosylation factor like GTPase 1 [Source:HGNC Symbol;Acc:HGNC:692] | 3957 | 0.405 | 0.4344 | Yes |
| 50 | DDIT4 | DNA damage inducible transcript 4 [Source:HGNC Symbol;Acc:HGNC:24944] | 3986 | 0.399 | 0.4371 | Yes |
| 51 | LAS1L | LAS1 like ribosome biogenesis factor [Source:HGNC Symbol;Acc:HGNC:25726] | 4071 | 0.385 | 0.4383 | Yes |
| 52 | IARS1 | isoleucyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:5330] | 4681 | 0.282 | 0.4243 | No |
| 53 | PDCD6 | programmed cell death 6 [Source:HGNC Symbol;Acc:HGNC:8765] | 4811 | 0.262 | 0.4231 | No |
| 54 | BIN3 | bridging integrator 3 [Source:HGNC Symbol;Acc:HGNC:1054] | 4812 | 0.262 | 0.4254 | No |
| 55 | ERCC3 | ERCC excision repair 3, TFIIH core complex helicase subunit [Source:HGNC Symbol;Acc:HGNC:3435] | 5479 | 0.162 | 0.4088 | No |
| 56 | PEX3 | peroxisomal biogenesis factor 3 [Source:HGNC Symbol;Acc:HGNC:8858] | 5514 | 0.158 | 0.4093 | No |
| 57 | NMI | N-myc and STAT interactor [Source:HGNC Symbol;Acc:HGNC:7854] | 5574 | 0.149 | 0.4090 | No |
| 58 | PEX19 | peroxisomal biogenesis factor 19 [Source:HGNC Symbol;Acc:HGNC:9713] | 5643 | 0.140 | 0.4084 | No |
| 59 | POLR1E | RNA polymerase I subunit E [Source:HGNC Symbol;Acc:HGNC:17631] | 5701 | 0.131 | 0.4080 | No |
| 60 | MOGS | mannosyl-oligosaccharide glucosidase [Source:HGNC Symbol;Acc:HGNC:24862] | 6140 | 0.075 | 0.3968 | No |
| 61 | RHOU | ras homolog family member U [Source:HGNC Symbol;Acc:HGNC:17794] | 6207 | 0.067 | 0.3956 | No |
| 62 | SAR1B | secretion associated Ras related GTPase 1B [Source:HGNC Symbol;Acc:HGNC:10535] | 6259 | 0.059 | 0.3947 | No |
| 63 | SRI | sorcin [Source:HGNC Symbol;Acc:HGNC:11292] | 6351 | 0.048 | 0.3927 | No |
| 64 | PTPN9 | protein tyrosine phosphatase non-receptor type 9 [Source:HGNC Symbol;Acc:HGNC:9661] | 6539 | 0.022 | 0.3878 | No |
| 65 | PRSS12 | serine protease 12 [Source:HGNC Symbol;Acc:HGNC:9477] | 6694 | 0.000 | 0.3837 | No |
| 66 | S100A9 | S100 calcium binding protein A9 [Source:HGNC Symbol;Acc:HGNC:10499] | 8830 | 0.000 | 0.3259 | No |
| 67 | S100A8 | S100 calcium binding protein A8 [Source:HGNC Symbol;Acc:HGNC:10498] | 8833 | 0.000 | 0.3259 | No |
| 68 | SLPI | secretory leukocyte peptidase inhibitor [Source:HGNC Symbol;Acc:HGNC:11092] | 9564 | 0.000 | 0.3061 | No |
| 69 | GSTA4 | glutathione S-transferase alpha 4 [Source:HGNC Symbol;Acc:HGNC:4629] | 10497 | 0.000 | 0.2809 | No |
| 70 | PPBP | pro-platelet basic protein [Source:HGNC Symbol;Acc:HGNC:9240] | 11212 | 0.000 | 0.2616 | No |
| 71 | C10orf99 | chromosome 10 open reading frame 99 [Source:HGNC Symbol;Acc:HGNC:31428] | 11369 | 0.000 | 0.2574 | No |
| 72 | PRND | prion like protein doppel [Source:HGNC Symbol;Acc:HGNC:15748] | 11795 | 0.000 | 0.2459 | No |
| 73 | S100A16 | S100 calcium binding protein A16 [Source:HGNC Symbol;Acc:HGNC:20441] | 11919 | 0.000 | 0.2425 | No |
| 74 | LPAR3 | lysophosphatidic acid receptor 3 [Source:HGNC Symbol;Acc:HGNC:14298] | 13924 | 0.000 | 0.1883 | No |
| 75 | CXCL12 | C-X-C motif chemokine ligand 12 [Source:HGNC Symbol;Acc:HGNC:10672] | 18907 | 0.000 | 0.0535 | No |
| 76 | GUCA2A | guanylate cyclase activator 2A [Source:HGNC Symbol;Acc:HGNC:4682] | 19215 | 0.000 | 0.0452 | No |
| 77 | GJB2 | gap junction protein beta 2 [Source:HGNC Symbol;Acc:HGNC:4284] | 20945 | 0.000 | -0.0015 | No |
| 78 | KRT79 | keratin 79 [Source:HGNC Symbol;Acc:HGNC:28930] | 22018 | 0.000 | -0.0305 | No |
| 79 | CTSL | cathepsin L [Source:HGNC Symbol;Acc:HGNC:2537] | 25093 | 0.000 | -0.1137 | No |
| 80 | STAT6 | signal transducer and activator of transcription 6 [Source:HGNC Symbol;Acc:HGNC:11368] | 28458 | -0.113 | -0.2037 | No |
| 81 | BDH1 | 3-hydroxybutyrate dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:1027] | 28746 | -0.149 | -0.2102 | No |
| 82 | MBNL3 | muscleblind like splicing regulator 3 [Source:HGNC Symbol;Acc:HGNC:20564] | 29559 | -0.247 | -0.2300 | No |
| 83 | ACADVL | acyl-CoA dehydrogenase very long chain [Source:HGNC Symbol;Acc:HGNC:92] | 29561 | -0.247 | -0.2278 | No |
| 84 | IDNK | IDNK gluconokinase [Source:HGNC Symbol;Acc:HGNC:31367] | 30086 | -0.316 | -0.2392 | No |
| 85 | SIGMAR1 | sigma non-opioid intracellular receptor 1 [Source:HGNC Symbol;Acc:HGNC:8157] | 30124 | -0.321 | -0.2374 | No |
| 86 | SYTL1 | synaptotagmin like 1 [Source:HGNC Symbol;Acc:HGNC:15584] | 30222 | -0.334 | -0.2371 | No |
| 87 | PIK3R4 | phosphoinositide-3-kinase regulatory subunit 4 [Source:HGNC Symbol;Acc:HGNC:8982] | 30320 | -0.344 | -0.2367 | No |
| 88 | NSUN2 | NOP2/Sun RNA methyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:25994] | 30345 | -0.347 | -0.2343 | No |
| 89 | COASY | Coenzyme A synthase [Source:HGNC Symbol;Acc:HGNC:29932] | 30594 | -0.380 | -0.2376 | No |
| 90 | ANXA4 | annexin A4 [Source:HGNC Symbol;Acc:HGNC:542] | 30670 | -0.391 | -0.2362 | No |
| 91 | TXNRD3 | thioredoxin reductase 3 [Source:HGNC Symbol;Acc:HGNC:20667] | 31096 | -0.454 | -0.2437 | No |
| 92 | PLEKHA7 | pleckstrin homology domain containing A7 [Source:HGNC Symbol;Acc:HGNC:27049] | 31132 | -0.458 | -0.2406 | No |
| 93 | AP3D1 | adaptor related protein complex 3 subunit delta 1 [Source:HGNC Symbol;Acc:HGNC:568] | 31207 | -0.469 | -0.2385 | No |
| 94 | DGAT1 | diacylglycerol O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:2843] | 31296 | -0.484 | -0.2366 | No |
| 95 | RDH11 | retinol dehydrogenase 11 [Source:HGNC Symbol;Acc:HGNC:17964] | 31310 | -0.486 | -0.2327 | No |
| 96 | EIF3C | eukaryotic translation initiation factor 3 subunit C [Source:HGNC Symbol;Acc:HGNC:3279] | 31522 | -0.526 | -0.2338 | No |
| 97 | POLR3G | RNA polymerase III subunit G [Source:HGNC Symbol;Acc:HGNC:30075] | 31641 | -0.547 | -0.2322 | No |
| 98 | PHF8 | PHD finger protein 8 [Source:HGNC Symbol;Acc:HGNC:20672] | 31648 | -0.548 | -0.2275 | No |
| 99 | COG6 | component of oligomeric golgi complex 6 [Source:HGNC Symbol;Acc:HGNC:18621] | 31650 | -0.548 | -0.2227 | No |
| 100 | RSRC1 | arginine and serine rich coiled-coil 1 [Source:HGNC Symbol;Acc:HGNC:24152] | 32148 | -0.642 | -0.2305 | No |
| 101 | CCDC90B | coiled-coil domain containing 90B [Source:HGNC Symbol;Acc:HGNC:28108] | 32359 | -0.684 | -0.2302 | No |
| 102 | UBE3B | ubiquitin protein ligase E3B [Source:HGNC Symbol;Acc:HGNC:13478] | 32397 | -0.693 | -0.2251 | No |
| 103 | STX7 | syntaxin 7 [Source:HGNC Symbol;Acc:HGNC:11442] | 32946 | -0.818 | -0.2327 | No |
| 104 | LPCAT3 | lysophosphatidylcholine acyltransferase 3 [Source:HGNC Symbol;Acc:HGNC:30244] | 32950 | -0.819 | -0.2256 | No |
| 105 | NUCB2 | nucleobindin 2 [Source:HGNC Symbol;Acc:HGNC:8044] | 32953 | -0.820 | -0.2184 | No |
| 106 | MTRES1 | mitochondrial transcription rescue factor 1 [Source:HGNC Symbol;Acc:HGNC:17971] | 32960 | -0.821 | -0.2114 | No |
| 107 | PPA2 | inorganic pyrophosphatase 2 [Source:HGNC Symbol;Acc:HGNC:28883] | 33333 | -0.922 | -0.2133 | No |
| 108 | CBR4 | carbonyl reductase 4 [Source:HGNC Symbol;Acc:HGNC:25891] | 33542 | -0.980 | -0.2103 | No |
| 109 | AIMP1 | aminoacyl tRNA synthetase complex interacting multifunctional protein 1 [Source:HGNC Symbol;Acc:HGNC:10648] | 33621 | -1.003 | -0.2036 | No |
| 110 | NME7 | NME/NM23 family member 7 [Source:HGNC Symbol;Acc:HGNC:20461] | 33636 | -1.009 | -0.1951 | No |
| 111 | AKAP9 | A-kinase anchoring protein 9 [Source:HGNC Symbol;Acc:HGNC:379] | 34016 | -1.134 | -0.1954 | No |
| 112 | ACP5 | acid phosphatase 5, tartrate resistant [Source:HGNC Symbol;Acc:HGNC:124] | 34130 | -1.173 | -0.1881 | No |
| 113 | TPRG1L | tumor protein p63 regulated 1 like [Source:HGNC Symbol;Acc:HGNC:27007] | 34292 | -1.230 | -0.1817 | No |
| 114 | HIGD2A | HIG1 hypoxia inducible domain family member 2A [Source:HGNC Symbol;Acc:HGNC:28311] | 34317 | -1.238 | -0.1714 | No |
| 115 | SPG11 | SPG11 vesicle trafficking associated, spatacsin [Source:HGNC Symbol;Acc:HGNC:11226] | 34771 | -1.426 | -0.1711 | No |
| 116 | LACTB2 | lactamase beta 2 [Source:HGNC Symbol;Acc:HGNC:18512] | 35399 | -1.726 | -0.1729 | No |
| 117 | H2BC4 | H2B clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4757] | 35513 | -1.779 | -0.1603 | No |
| 118 | COQ9 | coenzyme Q9 [Source:HGNC Symbol;Acc:HGNC:25302] | 35529 | -1.793 | -0.1450 | No |
| 119 | C3orf70 | chromosome 3 open reading frame 70 [Source:HGNC Symbol;Acc:HGNC:33731] | 35604 | -1.838 | -0.1308 | No |
| 120 | PANK1 | pantothenate kinase 1 [Source:HGNC Symbol;Acc:HGNC:8598] | 35816 | -1.986 | -0.1190 | No |
| 121 | GALNT11 | polypeptide N-acetylgalactosaminyltransferase 11 [Source:HGNC Symbol;Acc:HGNC:19875] | 36089 | -2.209 | -0.1069 | No |
| 122 | ELOVL6 | ELOVL fatty acid elongase 6 [Source:HGNC Symbol;Acc:HGNC:15829] | 36166 | -2.285 | -0.0889 | No |
| 123 | NIPSNAP1 | nipsnap homolog 1 [Source:HGNC Symbol;Acc:HGNC:7827] | 36601 | -2.781 | -0.0762 | No |
| 124 | PAFAH1B3 | platelet activating factor acetylhydrolase 1b catalytic subunit 3 [Source:HGNC Symbol;Acc:HGNC:8576] | 36761 | -3.086 | -0.0533 | No |
| 125 | GBE1 | 1,4-alpha-glucan branching enzyme 1 [Source:HGNC Symbol;Acc:HGNC:4180] | 36836 | -3.286 | -0.0264 | No |
| 126 | SCD | stearoyl-CoA desaturase [Source:HGNC Symbol;Acc:HGNC:10571] | 36973 | -3.782 | 0.0032 | No |