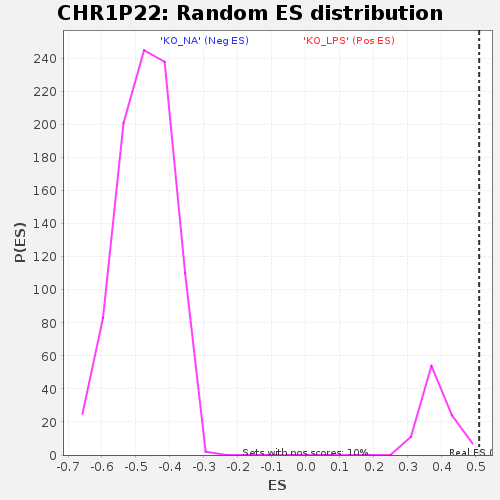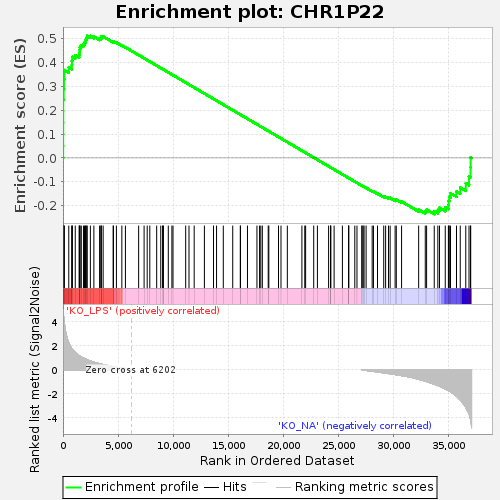
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_LPS_versus_KO_NA.greenbergcls.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_LPS |
| GeneSet | CHR1P22 |
| Enrichment Score (ES) | 0.51052475 |
| Normalized Enrichment Score (NES) | 1.32357 |
| Nominal p-value | 0.010416667 |
| FDR q-value | 0.35946417 |
| FWER p-Value | 0.752 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GBP7 | guanylate binding protein 7 [Source:HGNC Symbol;Acc:HGNC:29606] | 12 | 4.915 | 0.0492 | Yes |
| 2 | GBP3 | guanylate binding protein 3 [Source:HGNC Symbol;Acc:HGNC:4184] | 16 | 4.873 | 0.0982 | Yes |
| 3 | GBP6 | guanylate binding protein family member 6 [Source:HGNC Symbol;Acc:HGNC:25395] | 19 | 4.854 | 0.1470 | Yes |
| 4 | GBP1 | guanylate binding protein 1 [Source:HGNC Symbol;Acc:HGNC:4182] | 21 | 4.831 | 0.1957 | Yes |
| 5 | GBP5 | guanylate binding protein 5 [Source:HGNC Symbol;Acc:HGNC:19895] | 22 | 4.821 | 0.2442 | Yes |
| 6 | GBP1P1 | guanylate binding protein 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:39561] | 59 | 4.426 | 0.2878 | Yes |
| 7 | GBP2 | guanylate binding protein 2 [Source:HGNC Symbol;Acc:HGNC:4183] | 86 | 4.166 | 0.3291 | Yes |
| 8 | GBP4 | guanylate binding protein 4 [Source:HGNC Symbol;Acc:HGNC:20480] | 122 | 3.897 | 0.3674 | Yes |
| 9 | TMED5 | transmembrane p24 trafficking protein 5 [Source:HGNC Symbol;Acc:HGNC:24251] | 524 | 2.245 | 0.3792 | Yes |
| 10 | BCAR3 | BCAR3 adaptor protein, NSP family member [Source:HGNC Symbol;Acc:HGNC:973] | 803 | 1.745 | 0.3892 | Yes |
| 11 | ACTBP12 | ACTB pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:150] | 812 | 1.738 | 0.4065 | Yes |
| 12 | GCLM | glutamate-cysteine ligase modifier subunit [Source:HGNC Symbol;Acc:HGNC:4312] | 849 | 1.693 | 0.4226 | Yes |
| 13 | ZNHIT6 | zinc finger HIT-type containing 6 [Source:HGNC Symbol;Acc:HGNC:26089] | 1113 | 1.418 | 0.4298 | Yes |
| 14 | BCL10 | BCL10 immune signaling adaptor [Source:HGNC Symbol;Acc:HGNC:989] | 1462 | 1.167 | 0.4321 | Yes |
| 15 | MCOLN2 | mucolipin 2 [Source:HGNC Symbol;Acc:HGNC:13357] | 1513 | 1.137 | 0.4422 | Yes |
| 16 | LRRC8D | leucine rich repeat containing 8 VRAC subunit D [Source:HGNC Symbol;Acc:HGNC:16992] | 1521 | 1.134 | 0.4534 | Yes |
| 17 | FNBP1L | formin binding protein 1 like [Source:HGNC Symbol;Acc:HGNC:20851] | 1540 | 1.123 | 0.4643 | Yes |
| 18 | DNTTIP2 | deoxynucleotidyltransferase terminal interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:24013] | 1660 | 1.052 | 0.4716 | Yes |
| 19 | GTF2B | general transcription factor IIB [Source:HGNC Symbol;Acc:HGNC:4648] | 1858 | 0.961 | 0.4760 | Yes |
| 20 | DR1 | down-regulator of transcription 1 [Source:HGNC Symbol;Acc:HGNC:3017] | 1953 | 0.921 | 0.4827 | Yes |
| 21 | PRKAR1AP1 | protein kinase cAMP-dependent type I regulatory subunit alpha pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:9389] | 2046 | 0.882 | 0.4891 | Yes |
| 22 | MTF2 | metal response element binding transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:29535] | 2083 | 0.866 | 0.4969 | Yes |
| 23 | ZNF644 | zinc finger protein 644 [Source:HGNC Symbol;Acc:HGNC:29222] | 2156 | 0.836 | 0.5033 | Yes |
| 24 | GNG5 | G protein subunit gamma 5 [Source:HGNC Symbol;Acc:HGNC:4408] | 2195 | 0.816 | 0.5105 | Yes |
| 25 | PTGES3P1 | prostaglandin E synthase 3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:43824] | 2485 | 0.718 | 0.5099 | No |
| 26 | SH3GLB1 | SH3 domain containing GRB2 like, endophilin B1 [Source:HGNC Symbol;Acc:HGNC:10833] | 2803 | 0.618 | 0.5076 | No |
| 27 | RPF1 | ribosome production factor 1 homolog [Source:HGNC Symbol;Acc:HGNC:30350] | 3322 | 0.489 | 0.4985 | No |
| 28 | C1orf52 | chromosome 1 open reading frame 52 [Source:HGNC Symbol;Acc:HGNC:24871] | 3456 | 0.462 | 0.4996 | No |
| 29 | RBMXL1 | RBMX like 1 [Source:HGNC Symbol;Acc:HGNC:25073] | 3460 | 0.461 | 0.5041 | No |
| 30 | PKN2 | protein kinase N2 [Source:HGNC Symbol;Acc:HGNC:9406] | 3464 | 0.461 | 0.5087 | No |
| 31 | CAPNS1P1 | calpain small subunit 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:39562] | 3645 | 0.423 | 0.5081 | No |
| 32 | SELENOF | selenoprotein F [Source:HGNC Symbol;Acc:HGNC:17705] | 4550 | 0.255 | 0.4862 | No |
| 33 | WDR82P2 | WD repeat domain 82 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:33513] | 4583 | 0.249 | 0.4879 | No |
| 34 | SNORD21 | small nucleolar RNA, C/D box 21 [Source:HGNC Symbol;Acc:HGNC:10144] | 4848 | 0.207 | 0.4828 | No |
| 35 | RPL36AP10 | ribosomal protein L36a pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:36175] | 5342 | 0.132 | 0.4708 | No |
| 36 | ARHGAP29 | Rho GTPase activating protein 29 [Source:HGNC Symbol;Acc:HGNC:30207] | 5666 | 0.083 | 0.4629 | No |
| 37 | LINC01364 | long intergenic non-protein coding RNA 1364 [Source:HGNC Symbol;Acc:HGNC:50599] | 6866 | 0.000 | 0.4305 | No |
| 38 | HFM1 | helicase for meiosis 1 [Source:HGNC Symbol;Acc:HGNC:20193] | 7362 | 0.000 | 0.4171 | No |
| 39 | LINC01555 | long intergenic non-protein coding RNA 1555 [Source:HGNC Symbol;Acc:HGNC:26647] | 7643 | 0.000 | 0.4095 | No |
| 40 | RNU6-125P | RNA, U6 small nuclear 125, pseudogene [Source:HGNC Symbol;Acc:HGNC:47088] | 7879 | 0.000 | 0.4032 | No |
| 41 | LINC01140 | long intergenic non-protein coding RNA 1140 [Source:HGNC Symbol;Acc:HGNC:27922] | 8504 | 0.000 | 0.3863 | No |
| 42 | MTCO2P21 | MT-CO2 pseudogene 21 [Source:HGNC Symbol;Acc:HGNC:52150] | 8873 | 0.000 | 0.3763 | No |
| 43 | MIR4423 | microRNA 4423 [Source:HGNC Symbol;Acc:HGNC:41784] | 9040 | 0.000 | 0.3718 | No |
| 44 | MTATP6P13 | MT-ATP6 pseudogene 13 [Source:HGNC Symbol;Acc:HGNC:52058] | 9097 | 0.000 | 0.3703 | No |
| 45 | CHCHD2P5 | coiled-coil-helix-coiled-coil-helix domain containing 2 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:39589] | 9105 | 0.000 | 0.3701 | No |
| 46 | RN7SL692P | RNA, 7SL, cytoplasmic 692, pseudogene [Source:HGNC Symbol;Acc:HGNC:46708] | 9559 | 0.000 | 0.3579 | No |
| 47 | RN7SL440P | RNA, 7SL, cytoplasmic 440, pseudogene [Source:HGNC Symbol;Acc:HGNC:46456] | 9885 | 0.000 | 0.3491 | No |
| 48 | C1orf146 | chromosome 1 open reading frame 146 [Source:HGNC Symbol;Acc:HGNC:24032] | 9986 | 0.000 | 0.3464 | No |
| 49 | RN7SL653P | RNA, 7SL, cytoplasmic 653, pseudogene [Source:HGNC Symbol;Acc:HGNC:46669] | 11137 | 0.000 | 0.3153 | No |
| 50 | RNU4-59P | RNA, U4 small nuclear 59, pseudogene [Source:HGNC Symbol;Acc:HGNC:46995] | 11438 | 0.000 | 0.3072 | No |
| 51 | CCDC18-AS1 | CCDC18 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:52262] | 11909 | 0.000 | 0.2945 | No |
| 52 | HMGB3P9 | high mobility group box 3 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:39301] | 12836 | 0.000 | 0.2694 | No |
| 53 | LPAR3 | lysophosphatidic acid receptor 3 [Source:HGNC Symbol;Acc:HGNC:14298] | 13664 | 0.000 | 0.2470 | No |
| 54 | MTCO3P21 | MT-CO3 pseudogene 21 [Source:HGNC Symbol;Acc:HGNC:52083] | 13940 | 0.000 | 0.2396 | No |
| 55 | BARHL2 | BarH like homeobox 2 [Source:HGNC Symbol;Acc:HGNC:954] | 14551 | 0.000 | 0.2231 | No |
| 56 | RNU6-970P | RNA, U6 small nuclear 970, pseudogene [Source:HGNC Symbol;Acc:HGNC:47933] | 15408 | 0.000 | 0.2000 | No |
| 57 | RN7SL583P | RNA, 7SL, cytoplasmic 583, pseudogene [Source:HGNC Symbol;Acc:HGNC:46599] | 16094 | 0.000 | 0.1814 | No |
| 58 | SNORA66 | small nucleolar RNA, H/ACA box 66 [Source:HGNC Symbol;Acc:HGNC:10223] | 16114 | 0.000 | 0.1809 | No |
| 59 | PKN2-AS1 | PKN2 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:50597] | 16746 | 0.000 | 0.1639 | No |
| 60 | CCNJP2 | cyclin J pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:37997] | 17606 | 0.000 | 0.1406 | No |
| 61 | SYDE2 | synapse defective Rho GTPase homolog 2 [Source:HGNC Symbol;Acc:HGNC:25841] | 17843 | 0.000 | 0.1342 | No |
| 62 | RN7SKP272 | RN7SK pseudogene 272 [Source:HGNC Symbol;Acc:HGNC:45996] | 17955 | 0.000 | 0.1312 | No |
| 63 | RNU6-210P | RNA, U6 small nuclear 210, pseudogene [Source:HGNC Symbol;Acc:HGNC:47173] | 18096 | 0.000 | 0.1275 | No |
| 64 | DDAH1 | dimethylarginine dimethylaminohydrolase 1 [Source:HGNC Symbol;Acc:HGNC:2715] | 18628 | 0.000 | 0.1131 | No |
| 65 | MTND4P11 | MT-ND4 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:42223] | 18686 | 0.000 | 0.1115 | No |
| 66 | RNU6-695P | RNA, U6 small nuclear 695, pseudogene [Source:HGNC Symbol;Acc:HGNC:47658] | 19563 | 0.000 | 0.0879 | No |
| 67 | LINC01763 | long intergenic non-protein coding RNA 1763 [Source:HGNC Symbol;Acc:HGNC:52554] | 19789 | 0.000 | 0.0818 | No |
| 68 | RN7SKP123 | RN7SK pseudogene 123 [Source:HGNC Symbol;Acc:HGNC:45847] | 20365 | 0.000 | 0.0662 | No |
| 69 | CLCA3P | chloride channel accessory 3, pseudogene [Source:HGNC Symbol;Acc:HGNC:2017] | 21688 | 0.000 | 0.0305 | No |
| 70 | MIR760 | microRNA 760 [Source:HGNC Symbol;Acc:HGNC:33666] | 21948 | 0.000 | 0.0235 | No |
| 71 | RN7SL824P | RNA, 7SL, cytoplasmic 824, pseudogene [Source:HGNC Symbol;Acc:HGNC:46840] | 22025 | 0.000 | 0.0214 | No |
| 72 | CCN1 | cellular communication network factor 1 [Source:HGNC Symbol;Acc:HGNC:2654] | 22760 | 0.000 | 0.0016 | No |
| 73 | LPCAT2BP | lysophosphatidylcholine acyltransferase 2b, pseudogene [Source:HGNC Symbol;Acc:HGNC:38003] | 23095 | 0.000 | -0.0075 | No |
| 74 | MTND3P21 | MT-ND3 pseudogene 21 [Source:HGNC Symbol;Acc:HGNC:52167] | 24111 | 0.000 | -0.0349 | No |
| 75 | CLCA2 | chloride channel accessory 2 [Source:HGNC Symbol;Acc:HGNC:2016] | 24295 | 0.000 | -0.0399 | No |
| 76 | CLCA1 | chloride channel accessory 1 [Source:HGNC Symbol;Acc:HGNC:2015] | 24296 | 0.000 | -0.0399 | No |
| 77 | CLCA4 | chloride channel accessory 4 [Source:HGNC Symbol;Acc:HGNC:2018] | 24299 | 0.000 | -0.0399 | No |
| 78 | COL24A1 | collagen type XXIV alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:20821] | 24611 | 0.000 | -0.0483 | No |
| 79 | ELOCP19 | elongin C pseudogene 19 [Source:HGNC Symbol;Acc:HGNC:38155] | 25364 | 0.000 | -0.0687 | No |
| 80 | RNA5SP53 | RNA, 5S ribosomal pseudogene 53 [Source:HGNC Symbol;Acc:HGNC:42830] | 25929 | 0.000 | -0.0839 | No |
| 81 | RNA5SP52 | RNA, 5S ribosomal pseudogene 52 [Source:HGNC Symbol;Acc:HGNC:42829] | 25930 | 0.000 | -0.0839 | No |
| 82 | RNA5SP51 | RNA, 5S ribosomal pseudogene 51 [Source:HGNC Symbol;Acc:HGNC:42828] | 25932 | 0.000 | -0.0840 | No |
| 83 | ABCA4 | ATP binding cassette subfamily A member 4 [Source:HGNC Symbol;Acc:HGNC:34] | 26489 | 0.000 | -0.0990 | No |
| 84 | LRRC8C-DT | LRRC8C divergent transcript [Source:HGNC Symbol;Acc:HGNC:53731] | 26687 | 0.000 | -0.1043 | No |
| 85 | TGFBR3 | transforming growth factor beta receptor 3 [Source:HGNC Symbol;Acc:HGNC:11774] | 27131 | 0.000 | -0.1163 | No |
| 86 | SETSIP | SET like protein [Source:HGNC Symbol;Acc:HGNC:42937] | 27146 | -0.001 | -0.1167 | No |
| 87 | RPL5P6 | ribosomal protein L5 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:35890] | 27247 | -0.020 | -0.1192 | No |
| 88 | BRDT | bromodomain testis associated [Source:HGNC Symbol;Acc:HGNC:1105] | 27331 | -0.035 | -0.1211 | No |
| 89 | HSP90B3P | heat shock protein 90 beta family member 3, pseudogene [Source:HGNC Symbol;Acc:HGNC:12100] | 27515 | -0.060 | -0.1254 | No |
| 90 | MCOLN3 | mucolipin 3 [Source:HGNC Symbol;Acc:HGNC:13358] | 28066 | -0.125 | -0.1390 | No |
| 91 | EVI5 | ecotropic viral integration site 5 [Source:HGNC Symbol;Acc:HGNC:3501] | 28182 | -0.138 | -0.1408 | No |
| 92 | RPAP2 | RNA polymerase II associated protein 2 [Source:HGNC Symbol;Acc:HGNC:25791] | 28551 | -0.189 | -0.1488 | No |
| 93 | CDCA4P2 | cell division cycle associated 4 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:49771] | 29116 | -0.263 | -0.1614 | No |
| 94 | EPHX4 | epoxide hydrolase 4 [Source:HGNC Symbol;Acc:HGNC:23758] | 29288 | -0.286 | -0.1632 | No |
| 95 | KYAT3 | kynurenine aminotransferase 3 [Source:HGNC Symbol;Acc:HGNC:33238] | 29551 | -0.319 | -0.1670 | No |
| 96 | MTCO1P21 | MT-CO1 pseudogene 21 [Source:HGNC Symbol;Acc:HGNC:52086] | 29698 | -0.338 | -0.1676 | No |
| 97 | RPL5 | ribosomal protein L5 [Source:HGNC Symbol;Acc:HGNC:10360] | 30149 | -0.400 | -0.1757 | No |
| 98 | SPATA1 | spermatogenesis associated 1 [Source:HGNC Symbol;Acc:HGNC:14682] | 30257 | -0.416 | -0.1744 | No |
| 99 | LMO4 | LIM domain only 4 [Source:HGNC Symbol;Acc:HGNC:6644] | 30736 | -0.489 | -0.1824 | No |
| 100 | GFI1 | growth factor independent 1 transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:4237] | 32286 | -0.794 | -0.2163 | No |
| 101 | ODF2L | outer dense fiber of sperm tails 2 like [Source:HGNC Symbol;Acc:HGNC:29225] | 32886 | -0.951 | -0.2229 | No |
| 102 | ZNF326 | zinc finger protein 326 [Source:HGNC Symbol;Acc:HGNC:14104] | 33007 | -0.983 | -0.2163 | No |
| 103 | FEN1P1 | flap structure-specific endonuclease 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:3651] | 33701 | -1.212 | -0.2228 | No |
| 104 | CTBS | chitobiase [Source:HGNC Symbol;Acc:HGNC:2496] | 34027 | -1.319 | -0.2183 | No |
| 105 | SSX2IP | SSX family member 2 interacting protein [Source:HGNC Symbol;Acc:HGNC:16509] | 34176 | -1.381 | -0.2084 | No |
| 106 | GEMIN8P4 | gem nuclear organelle associated protein 8 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:37979] | 34701 | -1.592 | -0.2065 | No |
| 107 | GAPDHP46 | glyceraldehyde 3 phosphate dehydrogenase pseudogene 46 [Source:HGNC Symbol;Acc:HGNC:37804] | 34986 | -1.730 | -0.1968 | No |
| 108 | PHKA1P1 | phosphorylase kinase regulatory subunit alpha 1 pseudogene [Source:HGNC Symbol;Acc:HGNC:33919] | 35011 | -1.747 | -0.1799 | No |
| 109 | CDC7 | cell division cycle 7 [Source:HGNC Symbol;Acc:HGNC:1745] | 35063 | -1.780 | -0.1633 | No |
| 110 | BTBD8 | BTB domain containing 8 [Source:HGNC Symbol;Acc:HGNC:21019] | 35178 | -1.845 | -0.1478 | No |
| 111 | GLMN | glomulin, FKBP associated protein [Source:HGNC Symbol;Acc:HGNC:14373] | 35722 | -2.206 | -0.1403 | No |
| 112 | HS2ST1 | heparan sulfate 2-O-sulfotransferase 1 [Source:HGNC Symbol;Acc:HGNC:5193] | 36062 | -2.511 | -0.1241 | No |
| 113 | DIPK1A | divergent protein kinase domain 1A [Source:HGNC Symbol;Acc:HGNC:32213] | 36563 | -3.147 | -0.1060 | No |
| 114 | LRRC8B | leucine rich repeat containing 8 VRAC subunit B [Source:HGNC Symbol;Acc:HGNC:30692] | 36857 | -3.658 | -0.0770 | No |
| 115 | LRRC8C | leucine rich repeat containing 8 VRAC subunit C [Source:HGNC Symbol;Acc:HGNC:25075] | 37000 | -4.126 | -0.0393 | No |
| 116 | CCDC18 | coiled-coil domain containing 18 [Source:HGNC Symbol;Acc:HGNC:30370] | 37005 | -4.142 | 0.0023 | No |