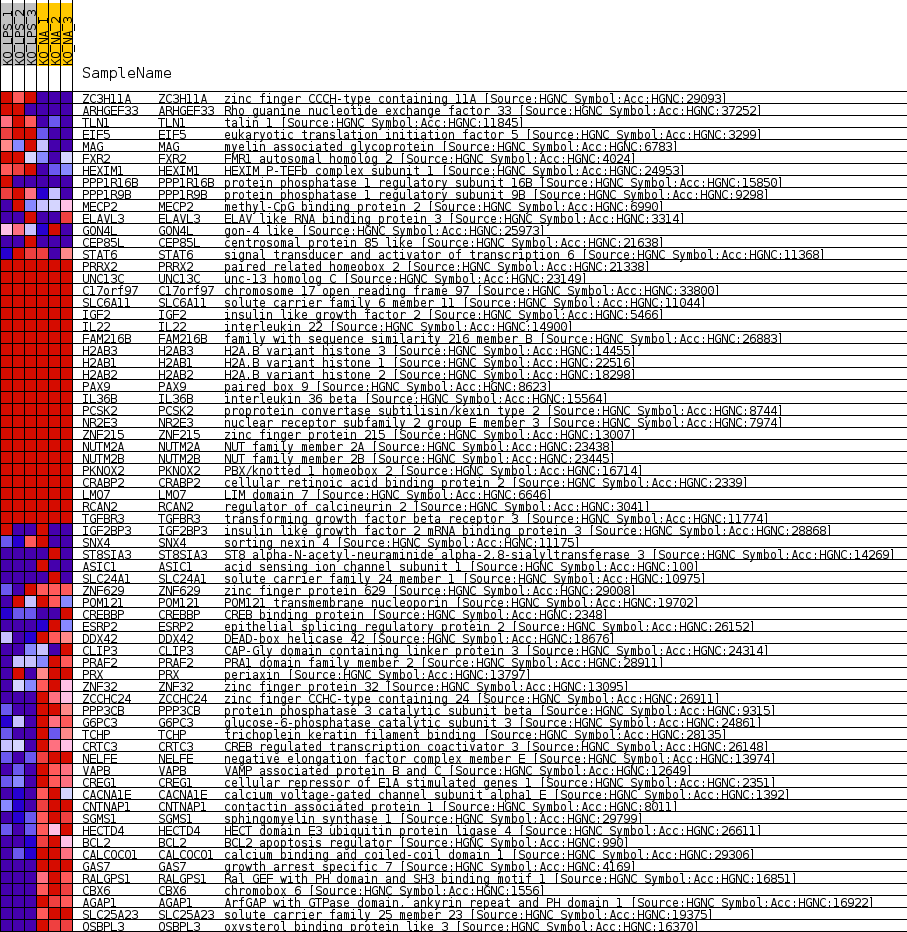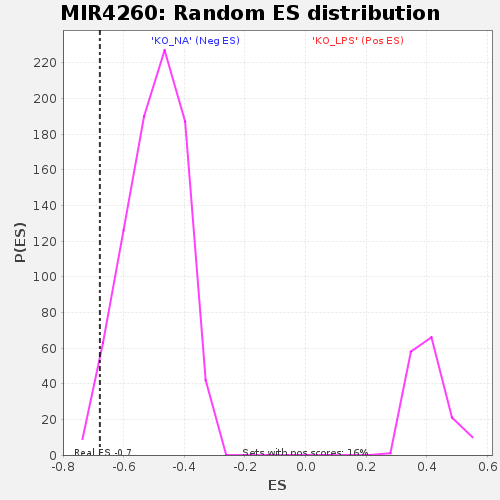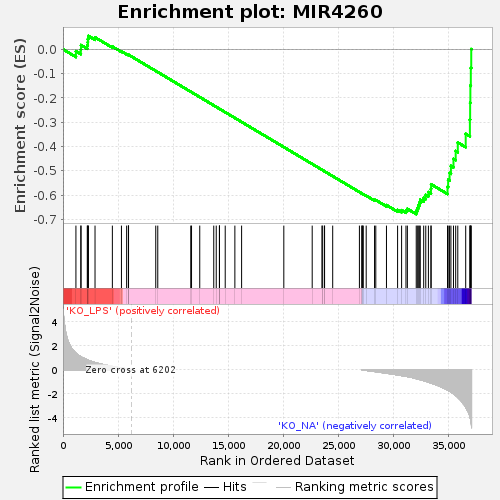
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_LPS_versus_KO_NA.greenbergcls.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR4260 |
| Enrichment Score (ES) | -0.6785741 |
| Normalized Enrichment Score (NES) | -1.3668303 |
| Nominal p-value | 0.030805686 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ZC3H11A | zinc finger CCCH-type containing 11A [Source:HGNC Symbol;Acc:HGNC:29093] | 1173 | 1.370 | -0.0077 | No |
| 2 | ARHGEF33 | Rho guanine nucleotide exchange factor 33 [Source:HGNC Symbol;Acc:HGNC:37252] | 1618 | 1.078 | -0.0008 | No |
| 3 | TLN1 | talin 1 [Source:HGNC Symbol;Acc:HGNC:11845] | 1637 | 1.067 | 0.0174 | No |
| 4 | EIF5 | eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] | 2204 | 0.814 | 0.0164 | No |
| 5 | MAG | myelin associated glycoprotein [Source:HGNC Symbol;Acc:HGNC:6783] | 2249 | 0.797 | 0.0292 | No |
| 6 | FXR2 | FMR1 autosomal homolog 2 [Source:HGNC Symbol;Acc:HGNC:4024] | 2261 | 0.794 | 0.0428 | No |
| 7 | HEXIM1 | HEXIM P-TEFb complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:24953] | 2308 | 0.776 | 0.0552 | No |
| 8 | PPP1R16B | protein phosphatase 1 regulatory subunit 16B [Source:HGNC Symbol;Acc:HGNC:15850] | 2907 | 0.586 | 0.0493 | No |
| 9 | PPP1R9B | protein phosphatase 1 regulatory subunit 9B [Source:HGNC Symbol;Acc:HGNC:9298] | 4481 | 0.265 | 0.0114 | No |
| 10 | MECP2 | methyl-CpG binding protein 2 [Source:HGNC Symbol;Acc:HGNC:6990] | 5305 | 0.135 | -0.0084 | No |
| 11 | ELAVL3 | ELAV like RNA binding protein 3 [Source:HGNC Symbol;Acc:HGNC:3314] | 5777 | 0.068 | -0.0200 | No |
| 12 | GON4L | gon-4 like [Source:HGNC Symbol;Acc:HGNC:25973] | 5939 | 0.043 | -0.0236 | No |
| 13 | CEP85L | centrosomal protein 85 like [Source:HGNC Symbol;Acc:HGNC:21638] | 5944 | 0.042 | -0.0229 | No |
| 14 | STAT6 | signal transducer and activator of transcription 6 [Source:HGNC Symbol;Acc:HGNC:11368] | 5950 | 0.041 | -0.0224 | No |
| 15 | PRRX2 | paired related homeobox 2 [Source:HGNC Symbol;Acc:HGNC:21338] | 8420 | 0.000 | -0.0890 | No |
| 16 | UNC13C | unc-13 homolog C [Source:HGNC Symbol;Acc:HGNC:23149] | 8603 | 0.000 | -0.0940 | No |
| 17 | C17orf97 | chromosome 17 open reading frame 97 [Source:HGNC Symbol;Acc:HGNC:33800] | 11598 | 0.000 | -0.1748 | No |
| 18 | SLC6A11 | solute carrier family 6 member 11 [Source:HGNC Symbol;Acc:HGNC:11044] | 11668 | 0.000 | -0.1767 | No |
| 19 | IGF2 | insulin like growth factor 2 [Source:HGNC Symbol;Acc:HGNC:5466] | 12414 | 0.000 | -0.1968 | No |
| 20 | IL22 | interleukin 22 [Source:HGNC Symbol;Acc:HGNC:14900] | 13685 | 0.000 | -0.2311 | No |
| 21 | FAM216B | family with sequence similarity 216 member B [Source:HGNC Symbol;Acc:HGNC:26883] | 13911 | 0.000 | -0.2372 | No |
| 22 | H2AB3 | H2A.B variant histone 3 [Source:HGNC Symbol;Acc:HGNC:14455] | 14202 | 0.000 | -0.2450 | No |
| 23 | H2AB1 | H2A.B variant histone 1 [Source:HGNC Symbol;Acc:HGNC:22516] | 14204 | 0.000 | -0.2451 | No |
| 24 | H2AB2 | H2A.B variant histone 2 [Source:HGNC Symbol;Acc:HGNC:18298] | 14205 | 0.000 | -0.2451 | No |
| 25 | PAX9 | paired box 9 [Source:HGNC Symbol;Acc:HGNC:8623] | 14726 | 0.000 | -0.2591 | No |
| 26 | IL36B | interleukin 36 beta [Source:HGNC Symbol;Acc:HGNC:15564] | 15602 | 0.000 | -0.2827 | No |
| 27 | PCSK2 | proprotein convertase subtilisin/kexin type 2 [Source:HGNC Symbol;Acc:HGNC:8744] | 16214 | 0.000 | -0.2992 | No |
| 28 | NR2E3 | nuclear receptor subfamily 2 group E member 3 [Source:HGNC Symbol;Acc:HGNC:7974] | 20045 | 0.000 | -0.4027 | No |
| 29 | ZNF215 | zinc finger protein 215 [Source:HGNC Symbol;Acc:HGNC:13007] | 22622 | 0.000 | -0.4723 | No |
| 30 | NUTM2A | NUT family member 2A [Source:HGNC Symbol;Acc:HGNC:23438] | 23519 | 0.000 | -0.4965 | No |
| 31 | NUTM2B | NUT family member 2B [Source:HGNC Symbol;Acc:HGNC:23445] | 23533 | 0.000 | -0.4968 | No |
| 32 | PKNOX2 | PBX/knotted 1 homeobox 2 [Source:HGNC Symbol;Acc:HGNC:16714] | 23721 | 0.000 | -0.5019 | No |
| 33 | CRABP2 | cellular retinoic acid binding protein 2 [Source:HGNC Symbol;Acc:HGNC:2339] | 23746 | 0.000 | -0.5025 | No |
| 34 | LMO7 | LIM domain 7 [Source:HGNC Symbol;Acc:HGNC:6646] | 24483 | 0.000 | -0.5224 | No |
| 35 | RCAN2 | regulator of calcineurin 2 [Source:HGNC Symbol;Acc:HGNC:3041] | 26908 | 0.000 | -0.5879 | No |
| 36 | TGFBR3 | transforming growth factor beta receptor 3 [Source:HGNC Symbol;Acc:HGNC:11774] | 27131 | 0.000 | -0.5939 | No |
| 37 | IGF2BP3 | insulin like growth factor 2 mRNA binding protein 3 [Source:HGNC Symbol;Acc:HGNC:28868] | 27143 | -0.000 | -0.5942 | No |
| 38 | SNX4 | sorting nexin 4 [Source:HGNC Symbol;Acc:HGNC:11175] | 27201 | -0.010 | -0.5956 | No |
| 39 | ST8SIA3 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 [Source:HGNC Symbol;Acc:HGNC:14269] | 27256 | -0.021 | -0.5966 | No |
| 40 | ASIC1 | acid sensing ion channel subunit 1 [Source:HGNC Symbol;Acc:HGNC:100] | 27524 | -0.061 | -0.6028 | No |
| 41 | SLC24A1 | solute carrier family 24 member 1 [Source:HGNC Symbol;Acc:HGNC:10975] | 28281 | -0.152 | -0.6205 | No |
| 42 | ZNF629 | zinc finger protein 629 [Source:HGNC Symbol;Acc:HGNC:29008] | 28395 | -0.167 | -0.6207 | No |
| 43 | POM121 | POM121 transmembrane nucleoporin [Source:HGNC Symbol;Acc:HGNC:19702] | 29364 | -0.296 | -0.6416 | No |
| 44 | CREBBP | CREB binding protein [Source:HGNC Symbol;Acc:HGNC:2348] | 30374 | -0.433 | -0.6613 | No |
| 45 | ESRP2 | epithelial splicing regulatory protein 2 [Source:HGNC Symbol;Acc:HGNC:26152] | 30738 | -0.489 | -0.6625 | No |
| 46 | DDX42 | DEAD-box helicase 42 [Source:HGNC Symbol;Acc:HGNC:18676] | 31134 | -0.553 | -0.6635 | No |
| 47 | CLIP3 | CAP-Gly domain containing linker protein 3 [Source:HGNC Symbol;Acc:HGNC:24314] | 31262 | -0.574 | -0.6569 | No |
| 48 | PRAF2 | PRA1 domain family member 2 [Source:HGNC Symbol;Acc:HGNC:28911] | 32066 | -0.747 | -0.6655 | Yes |
| 49 | PRX | periaxin [Source:HGNC Symbol;Acc:HGNC:13797] | 32161 | -0.769 | -0.6546 | Yes |
| 50 | ZNF32 | zinc finger protein 32 [Source:HGNC Symbol;Acc:HGNC:13095] | 32252 | -0.786 | -0.6432 | Yes |
| 51 | ZCCHC24 | zinc finger CCHC-type containing 24 [Source:HGNC Symbol;Acc:HGNC:26911] | 32346 | -0.810 | -0.6315 | Yes |
| 52 | PPP3CB | protein phosphatase 3 catalytic subunit beta [Source:HGNC Symbol;Acc:HGNC:9315] | 32433 | -0.833 | -0.6192 | Yes |
| 53 | G6PC3 | glucose-6-phosphatase catalytic subunit 3 [Source:HGNC Symbol;Acc:HGNC:24861] | 32743 | -0.914 | -0.6116 | Yes |
| 54 | TCHP | trichoplein keratin filament binding [Source:HGNC Symbol;Acc:HGNC:28135] | 32932 | -0.965 | -0.5997 | Yes |
| 55 | CRTC3 | CREB regulated transcription coactivator 3 [Source:HGNC Symbol;Acc:HGNC:26148] | 33171 | -1.035 | -0.5880 | Yes |
| 56 | NELFE | negative elongation factor complex member E [Source:HGNC Symbol;Acc:HGNC:13974] | 33400 | -1.109 | -0.5748 | Yes |
| 57 | VAPB | VAMP associated protein B and C [Source:HGNC Symbol;Acc:HGNC:12649] | 33418 | -1.114 | -0.5557 | Yes |
| 58 | CREG1 | cellular repressor of E1A stimulated genes 1 [Source:HGNC Symbol;Acc:HGNC:2351] | 34905 | -1.690 | -0.5662 | Yes |
| 59 | CACNA1E | calcium voltage-gated channel subunit alpha1 E [Source:HGNC Symbol;Acc:HGNC:1392] | 34963 | -1.719 | -0.5376 | Yes |
| 60 | CNTNAP1 | contactin associated protein 1 [Source:HGNC Symbol;Acc:HGNC:8011] | 35092 | -1.797 | -0.5096 | Yes |
| 61 | SGMS1 | sphingomyelin synthase 1 [Source:HGNC Symbol;Acc:HGNC:29799] | 35215 | -1.868 | -0.4802 | Yes |
| 62 | HECTD4 | HECT domain E3 ubiquitin protein ligase 4 [Source:HGNC Symbol;Acc:HGNC:26611] | 35447 | -2.011 | -0.4512 | Yes |
| 63 | BCL2 | BCL2 apoptosis regulator [Source:HGNC Symbol;Acc:HGNC:990] | 35648 | -2.151 | -0.4189 | Yes |
| 64 | CALCOCO1 | calcium binding and coiled-coil domain 1 [Source:HGNC Symbol;Acc:HGNC:29306] | 35839 | -2.300 | -0.3837 | Yes |
| 65 | GAS7 | growth arrest specific 7 [Source:HGNC Symbol;Acc:HGNC:4169] | 36557 | -3.135 | -0.3481 | Yes |
| 66 | RALGPS1 | Ral GEF with PH domain and SH3 binding motif 1 [Source:HGNC Symbol;Acc:HGNC:16851] | 36937 | -3.923 | -0.2896 | Yes |
| 67 | CBX6 | chromobox 6 [Source:HGNC Symbol;Acc:HGNC:1556] | 36960 | -4.003 | -0.2201 | Yes |
| 68 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 [Source:HGNC Symbol;Acc:HGNC:16922] | 36981 | -4.058 | -0.1495 | Yes |
| 69 | SLC25A23 | solute carrier family 25 member 23 [Source:HGNC Symbol;Acc:HGNC:19375] | 37021 | -4.211 | -0.0767 | Yes |
| 70 | OSBPL3 | oxysterol binding protein like 3 [Source:HGNC Symbol;Acc:HGNC:16370] | 37060 | -4.484 | 0.0008 | Yes |