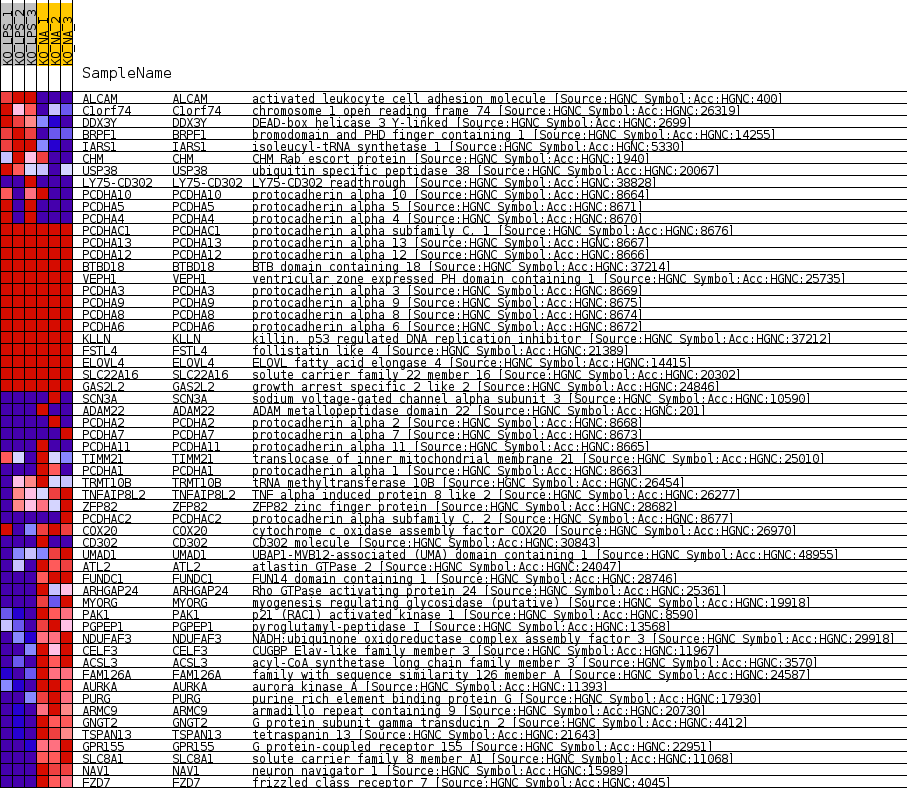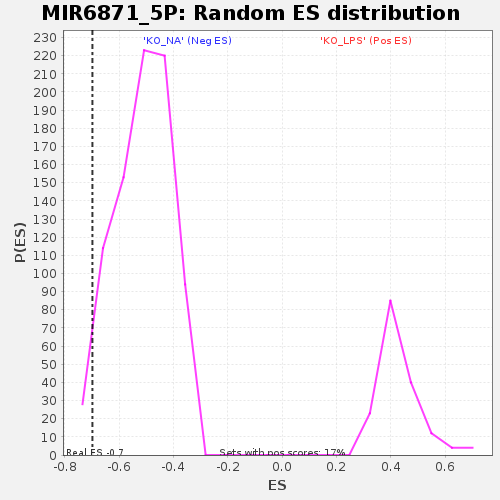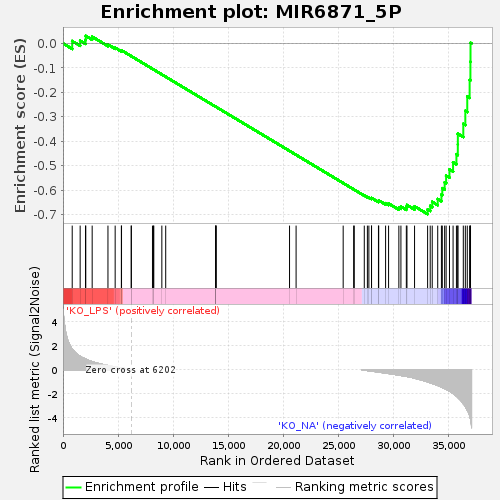
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_LPS_versus_KO_NA.greenbergcls.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR6871_5P |
| Enrichment Score (ES) | -0.69813955 |
| Normalized Enrichment Score (NES) | -1.356139 |
| Nominal p-value | 0.03125 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ALCAM | activated leukocyte cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:400] | 836 | 1.712 | 0.0098 | No |
| 2 | C1orf74 | chromosome 1 open reading frame 74 [Source:HGNC Symbol;Acc:HGNC:26319] | 1557 | 1.113 | 0.0114 | No |
| 3 | DDX3Y | DEAD-box helicase 3 Y-linked [Source:HGNC Symbol;Acc:HGNC:2699] | 2042 | 0.882 | 0.0150 | No |
| 4 | BRPF1 | bromodomain and PHD finger containing 1 [Source:HGNC Symbol;Acc:HGNC:14255] | 2068 | 0.873 | 0.0308 | No |
| 5 | IARS1 | isoleucyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:5330] | 2648 | 0.665 | 0.0277 | No |
| 6 | CHM | CHM Rab escort protein [Source:HGNC Symbol;Acc:HGNC:1940] | 4081 | 0.344 | -0.0044 | No |
| 7 | USP38 | ubiquitin specific peptidase 38 [Source:HGNC Symbol;Acc:HGNC:20067] | 4735 | 0.226 | -0.0178 | No |
| 8 | LY75-CD302 | LY75-CD302 readthrough [Source:HGNC Symbol;Acc:HGNC:38828] | 5298 | 0.136 | -0.0304 | No |
| 9 | PCDHA10 | protocadherin alpha 10 [Source:HGNC Symbol;Acc:HGNC:8664] | 5313 | 0.134 | -0.0283 | No |
| 10 | PCDHA5 | protocadherin alpha 5 [Source:HGNC Symbol;Acc:HGNC:8671] | 6198 | 0.000 | -0.0522 | No |
| 11 | PCDHA4 | protocadherin alpha 4 [Source:HGNC Symbol;Acc:HGNC:8670] | 6199 | 0.000 | -0.0521 | No |
| 12 | PCDHAC1 | protocadherin alpha subfamily C, 1 [Source:HGNC Symbol;Acc:HGNC:8676] | 8122 | 0.000 | -0.1040 | No |
| 13 | PCDHA13 | protocadherin alpha 13 [Source:HGNC Symbol;Acc:HGNC:8667] | 8229 | 0.000 | -0.1069 | No |
| 14 | PCDHA12 | protocadherin alpha 12 [Source:HGNC Symbol;Acc:HGNC:8666] | 8231 | 0.000 | -0.1069 | No |
| 15 | BTBD18 | BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:37214] | 8973 | 0.000 | -0.1269 | No |
| 16 | VEPH1 | ventricular zone expressed PH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25735] | 9324 | 0.000 | -0.1364 | No |
| 17 | PCDHA3 | protocadherin alpha 3 [Source:HGNC Symbol;Acc:HGNC:8669] | 13875 | 0.000 | -0.2593 | No |
| 18 | PCDHA9 | protocadherin alpha 9 [Source:HGNC Symbol;Acc:HGNC:8675] | 13876 | 0.000 | -0.2593 | No |
| 19 | PCDHA8 | protocadherin alpha 8 [Source:HGNC Symbol;Acc:HGNC:8674] | 13877 | 0.000 | -0.2593 | No |
| 20 | PCDHA6 | protocadherin alpha 6 [Source:HGNC Symbol;Acc:HGNC:8672] | 13878 | 0.000 | -0.2593 | No |
| 21 | KLLN | killin, p53 regulated DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:37212] | 20565 | 0.000 | -0.4398 | No |
| 22 | FSTL4 | follistatin like 4 [Source:HGNC Symbol;Acc:HGNC:21389] | 21160 | 0.000 | -0.4558 | No |
| 23 | ELOVL4 | ELOVL fatty acid elongase 4 [Source:HGNC Symbol;Acc:HGNC:14415] | 25434 | 0.000 | -0.5712 | No |
| 24 | SLC22A16 | solute carrier family 22 member 16 [Source:HGNC Symbol;Acc:HGNC:20302] | 26386 | 0.000 | -0.5969 | No |
| 25 | GAS2L2 | growth arrest specific 2 like 2 [Source:HGNC Symbol;Acc:HGNC:24846] | 26440 | 0.000 | -0.5983 | No |
| 26 | SCN3A | sodium voltage-gated channel alpha subunit 3 [Source:HGNC Symbol;Acc:HGNC:10590] | 27350 | -0.038 | -0.6222 | No |
| 27 | ADAM22 | ADAM metallopeptidase domain 22 [Source:HGNC Symbol;Acc:HGNC:201] | 27632 | -0.074 | -0.6284 | No |
| 28 | PCDHA2 | protocadherin alpha 2 [Source:HGNC Symbol;Acc:HGNC:8668] | 27767 | -0.090 | -0.6303 | No |
| 29 | PCDHA7 | protocadherin alpha 7 [Source:HGNC Symbol;Acc:HGNC:8673] | 28005 | -0.119 | -0.6344 | No |
| 30 | PCDHA11 | protocadherin alpha 11 [Source:HGNC Symbol;Acc:HGNC:8665] | 28011 | -0.120 | -0.6323 | No |
| 31 | TIMM21 | translocase of inner mitochondrial membrane 21 [Source:HGNC Symbol;Acc:HGNC:25010] | 28637 | -0.198 | -0.6454 | No |
| 32 | PCDHA1 | protocadherin alpha 1 [Source:HGNC Symbol;Acc:HGNC:8663] | 28660 | -0.203 | -0.6422 | No |
| 33 | TRMT10B | tRNA methyltransferase 10B [Source:HGNC Symbol;Acc:HGNC:26454] | 29285 | -0.286 | -0.6536 | No |
| 34 | TNFAIP8L2 | TNF alpha induced protein 8 like 2 [Source:HGNC Symbol;Acc:HGNC:26277] | 29556 | -0.319 | -0.6549 | No |
| 35 | ZFP82 | ZFP82 zinc finger protein [Source:HGNC Symbol;Acc:HGNC:28682] | 30489 | -0.451 | -0.6715 | No |
| 36 | PCDHAC2 | protocadherin alpha subfamily C, 2 [Source:HGNC Symbol;Acc:HGNC:8677] | 30676 | -0.480 | -0.6675 | No |
| 37 | COX20 | cytochrome c oxidase assembly factor COX20 [Source:HGNC Symbol;Acc:HGNC:26970] | 31167 | -0.560 | -0.6702 | No |
| 38 | CD302 | CD302 molecule [Source:HGNC Symbol;Acc:HGNC:30843] | 31225 | -0.569 | -0.6610 | No |
| 39 | UMAD1 | UBAP1-MVB12-associated (UMA) domain containing 1 [Source:HGNC Symbol;Acc:HGNC:48955] | 31914 | -0.707 | -0.6662 | No |
| 40 | ATL2 | atlastin GTPase 2 [Source:HGNC Symbol;Acc:HGNC:24047] | 33099 | -1.012 | -0.6790 | Yes |
| 41 | FUNDC1 | FUN14 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28746] | 33335 | -1.088 | -0.6648 | Yes |
| 42 | ARHGAP24 | Rho GTPase activating protein 24 [Source:HGNC Symbol;Acc:HGNC:25361] | 33518 | -1.147 | -0.6480 | Yes |
| 43 | MYORG | myogenesis regulating glycosidase (putative) [Source:HGNC Symbol;Acc:HGNC:19918] | 34017 | -1.316 | -0.6366 | Yes |
| 44 | PAK1 | p21 (RAC1) activated kinase 1 [Source:HGNC Symbol;Acc:HGNC:8590] | 34353 | -1.455 | -0.6182 | Yes |
| 45 | PGPEP1 | pyroglutamyl-peptidase I [Source:HGNC Symbol;Acc:HGNC:13568] | 34432 | -1.486 | -0.5922 | Yes |
| 46 | NDUFAF3 | NADH:ubiquinone oxidoreductase complex assembly factor 3 [Source:HGNC Symbol;Acc:HGNC:29918] | 34646 | -1.573 | -0.5682 | Yes |
| 47 | CELF3 | CUGBP Elav-like family member 3 [Source:HGNC Symbol;Acc:HGNC:11967] | 34780 | -1.632 | -0.5410 | Yes |
| 48 | ACSL3 | acyl-CoA synthetase long chain family member 3 [Source:HGNC Symbol;Acc:HGNC:3570] | 35084 | -1.792 | -0.5153 | Yes |
| 49 | FAM126A | family with sequence similarity 126 member A [Source:HGNC Symbol;Acc:HGNC:24587] | 35411 | -1.986 | -0.4866 | Yes |
| 50 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35715 | -2.202 | -0.4532 | Yes |
| 51 | PURG | purine rich element binding protein G [Source:HGNC Symbol;Acc:HGNC:17930] | 35841 | -2.302 | -0.4130 | Yes |
| 52 | ARMC9 | armadillo repeat containing 9 [Source:HGNC Symbol;Acc:HGNC:20730] | 35844 | -2.311 | -0.3694 | Yes |
| 53 | GNGT2 | G protein subunit gamma transducin 2 [Source:HGNC Symbol;Acc:HGNC:4412] | 36339 | -2.844 | -0.3290 | Yes |
| 54 | TSPAN13 | tetraspanin 13 [Source:HGNC Symbol;Acc:HGNC:21643] | 36523 | -3.071 | -0.2759 | Yes |
| 55 | GPR155 | G protein-coupled receptor 155 [Source:HGNC Symbol;Acc:HGNC:22951] | 36699 | -3.373 | -0.2169 | Yes |
| 56 | SLC8A1 | solute carrier family 8 member A1 [Source:HGNC Symbol;Acc:HGNC:11068] | 36914 | -3.851 | -0.1500 | Yes |
| 57 | NAV1 | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | 36985 | -4.076 | -0.0748 | Yes |
| 58 | FZD7 | frizzled class receptor 7 [Source:HGNC Symbol;Acc:HGNC:4045] | 36994 | -4.109 | 0.0026 | Yes |