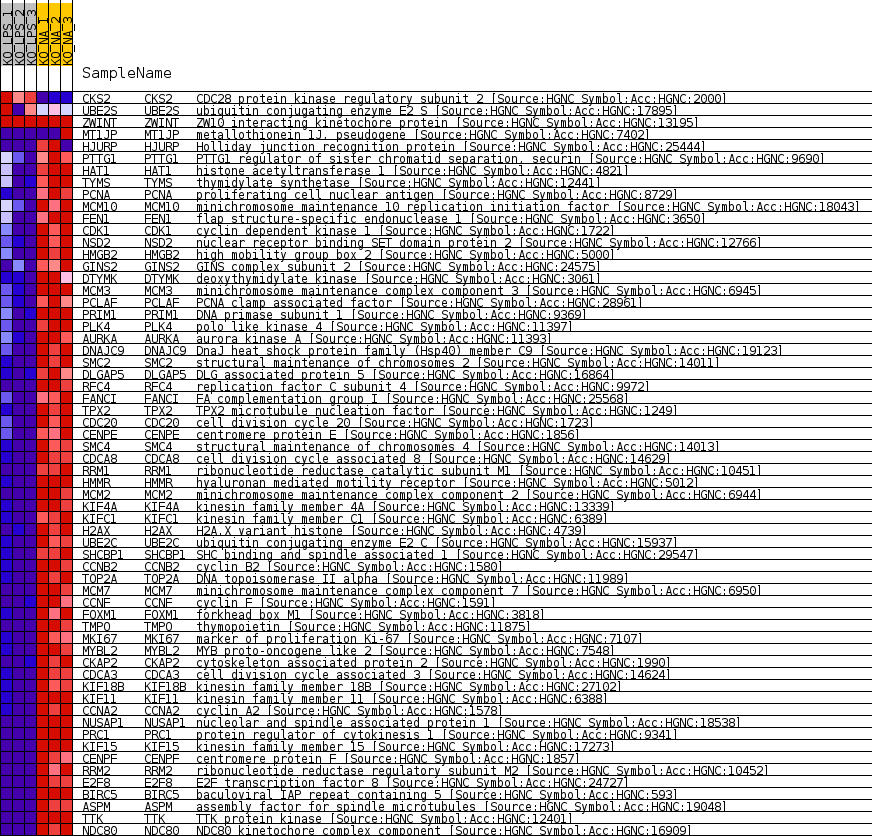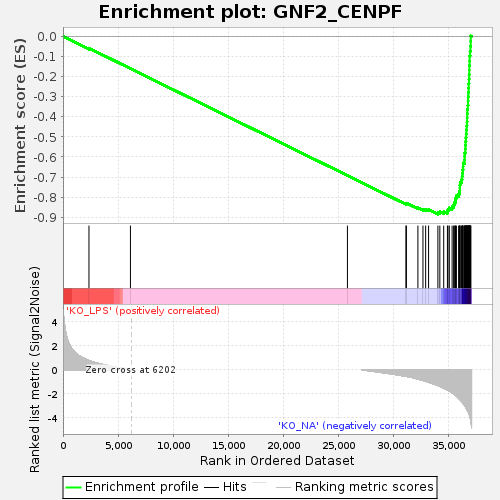
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_LPS_versus_KO_NA.greenbergcls.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_CENPF |
| Enrichment Score (ES) | -0.88422316 |
| Normalized Enrichment Score (NES) | -1.7472957 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 2357 | 0.761 | -0.0589 | No |
| 2 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 6122 | 0.013 | -0.1605 | No |
| 3 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 25820 | 0.000 | -0.6924 | No |
| 4 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 31130 | -0.553 | -0.8324 | No |
| 5 | HJURP | Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] | 31175 | -0.560 | -0.8301 | No |
| 6 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 32217 | -0.779 | -0.8533 | No |
| 7 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 32676 | -0.898 | -0.8601 | No |
| 8 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32921 | -0.962 | -0.8607 | No |
| 9 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33187 | -1.040 | -0.8614 | No |
| 10 | MCM10 | minichromosome maintenance 10 replication initiation factor [Source:HGNC Symbol;Acc:HGNC:18043] | 34033 | -1.321 | -0.8760 | Yes |
| 11 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 34209 | -1.394 | -0.8721 | Yes |
| 12 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 34560 | -1.537 | -0.8720 | Yes |
| 13 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 34898 | -1.687 | -0.8706 | Yes |
| 14 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34948 | -1.710 | -0.8612 | Yes |
| 15 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35070 | -1.784 | -0.8534 | Yes |
| 16 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 35322 | -1.930 | -0.8482 | Yes |
| 17 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35462 | -2.024 | -0.8394 | Yes |
| 18 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35526 | -2.066 | -0.8282 | Yes |
| 19 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35613 | -2.129 | -0.8173 | Yes |
| 20 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 35629 | -2.136 | -0.8044 | Yes |
| 21 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35715 | -2.202 | -0.7930 | Yes |
| 22 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 35912 | -2.379 | -0.7835 | Yes |
| 23 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35996 | -2.449 | -0.7705 | Yes |
| 24 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 36021 | -2.474 | -0.7558 | Yes |
| 25 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36023 | -2.475 | -0.7404 | Yes |
| 26 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 36050 | -2.499 | -0.7255 | Yes |
| 27 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 36181 | -2.640 | -0.7126 | Yes |
| 28 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 36232 | -2.692 | -0.6972 | Yes |
| 29 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 36254 | -2.721 | -0.6809 | Yes |
| 30 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 36278 | -2.752 | -0.6644 | Yes |
| 31 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 36318 | -2.818 | -0.6479 | Yes |
| 32 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36325 | -2.825 | -0.6305 | Yes |
| 33 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36448 | -2.956 | -0.6154 | Yes |
| 34 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36458 | -2.975 | -0.5971 | Yes |
| 35 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 36465 | -2.988 | -0.5787 | Yes |
| 36 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 36527 | -3.075 | -0.5612 | Yes |
| 37 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36533 | -3.091 | -0.5421 | Yes |
| 38 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 36549 | -3.110 | -0.5232 | Yes |
| 39 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 36578 | -3.169 | -0.5042 | Yes |
| 40 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36597 | -3.196 | -0.4849 | Yes |
| 41 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36619 | -3.233 | -0.4653 | Yes |
| 42 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36647 | -3.278 | -0.4456 | Yes |
| 43 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 36674 | -3.329 | -0.4256 | Yes |
| 44 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36684 | -3.340 | -0.4051 | Yes |
| 45 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36690 | -3.355 | -0.3844 | Yes |
| 46 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36710 | -3.387 | -0.3638 | Yes |
| 47 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36749 | -3.440 | -0.3435 | Yes |
| 48 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 36771 | -3.483 | -0.3224 | Yes |
| 49 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36778 | -3.498 | -0.3008 | Yes |
| 50 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36802 | -3.541 | -0.2794 | Yes |
| 51 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36819 | -3.570 | -0.2576 | Yes |
| 52 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36831 | -3.600 | -0.2355 | Yes |
| 53 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36856 | -3.657 | -0.2134 | Yes |
| 54 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36868 | -3.678 | -0.1908 | Yes |
| 55 | KIF15 | kinesin family member 15 [Source:HGNC Symbol;Acc:HGNC:17273] | 36883 | -3.727 | -0.1680 | Yes |
| 56 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36886 | -3.735 | -0.1448 | Yes |
| 57 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36911 | -3.844 | -0.1216 | Yes |
| 58 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36915 | -3.854 | -0.0977 | Yes |
| 59 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36957 | -3.986 | -0.0740 | Yes |
| 60 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36988 | -4.101 | -0.0493 | Yes |
| 61 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 37012 | -4.172 | -0.0239 | Yes |
| 62 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 37016 | -4.184 | 0.0020 | Yes |