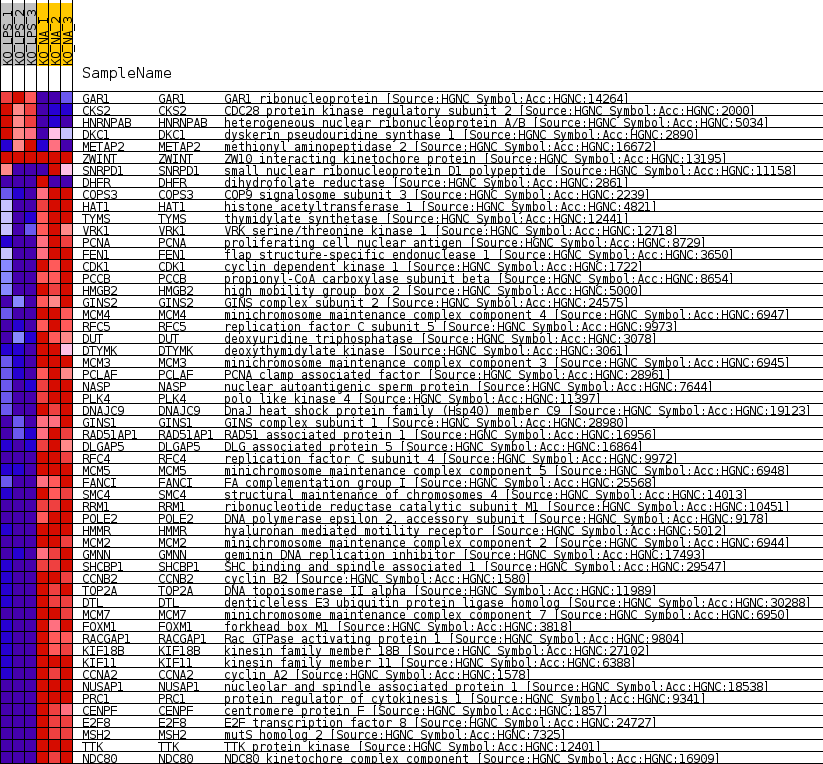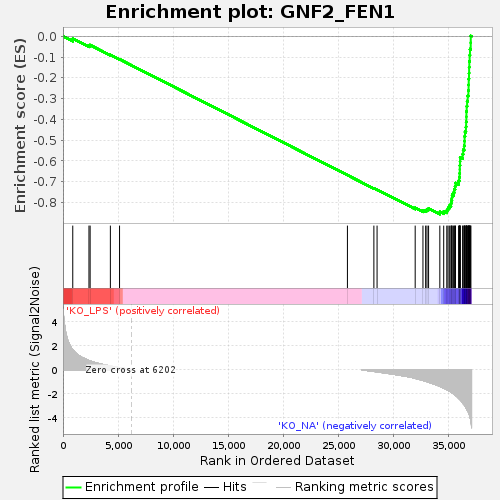
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #KO_LPS_versus_KO_NA.greenbergcls.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | greenbergcls.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_FEN1 |
| Enrichment Score (ES) | -0.8566021 |
| Normalized Enrichment Score (NES) | -1.6567175 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.1766526E-4 |
| FWER p-Value | 0.004 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GAR1 | GAR1 ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:14264] | 886 | 1.656 | -0.0110 | No |
| 2 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 2357 | 0.761 | -0.0447 | No |
| 3 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B [Source:HGNC Symbol;Acc:HGNC:5034] | 2463 | 0.725 | -0.0419 | No |
| 4 | DKC1 | dyskerin pseudouridine synthase 1 [Source:HGNC Symbol;Acc:HGNC:2890] | 4300 | 0.299 | -0.0892 | No |
| 5 | METAP2 | methionyl aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:16672] | 5133 | 0.163 | -0.1103 | No |
| 6 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 25820 | 0.000 | -0.6689 | No |
| 7 | SNRPD1 | small nuclear ribonucleoprotein D1 polypeptide [Source:HGNC Symbol;Acc:HGNC:11158] | 28221 | -0.143 | -0.7326 | No |
| 8 | DHFR | dihydrofolate reductase [Source:HGNC Symbol;Acc:HGNC:2861] | 28511 | -0.184 | -0.7390 | No |
| 9 | COPS3 | COP9 signalosome subunit 3 [Source:HGNC Symbol;Acc:HGNC:2239] | 31971 | -0.721 | -0.8267 | No |
| 10 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 32676 | -0.898 | -0.8387 | No |
| 11 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32921 | -0.962 | -0.8378 | No |
| 12 | VRK1 | VRK serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:12718] | 33043 | -0.994 | -0.8333 | No |
| 13 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33187 | -1.040 | -0.8290 | No |
| 14 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 34209 | -1.394 | -0.8457 | Yes |
| 15 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 34560 | -1.537 | -0.8432 | Yes |
| 16 | PCCB | propionyl-CoA carboxylase subunit beta [Source:HGNC Symbol;Acc:HGNC:8654] | 34841 | -1.659 | -0.8378 | Yes |
| 17 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34948 | -1.710 | -0.8273 | Yes |
| 18 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35070 | -1.784 | -0.8166 | Yes |
| 19 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 35218 | -1.870 | -0.8060 | Yes |
| 20 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 35250 | -1.890 | -0.7921 | Yes |
| 21 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 35306 | -1.922 | -0.7785 | Yes |
| 22 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 35322 | -1.930 | -0.7639 | Yes |
| 23 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35462 | -2.024 | -0.7518 | Yes |
| 24 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35526 | -2.066 | -0.7374 | Yes |
| 25 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35590 | -2.112 | -0.7226 | Yes |
| 26 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 35629 | -2.136 | -0.7070 | Yes |
| 27 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 35912 | -2.379 | -0.6960 | Yes |
| 28 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 35976 | -2.431 | -0.6787 | Yes |
| 29 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 36006 | -2.459 | -0.6603 | Yes |
| 30 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 36021 | -2.474 | -0.6414 | Yes |
| 31 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36023 | -2.475 | -0.6221 | Yes |
| 32 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 36043 | -2.495 | -0.6031 | Yes |
| 33 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 36050 | -2.499 | -0.5837 | Yes |
| 34 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 36278 | -2.752 | -0.5684 | Yes |
| 35 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36325 | -2.825 | -0.5476 | Yes |
| 36 | POLE2 | DNA polymerase epsilon 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9178] | 36416 | -2.915 | -0.5272 | Yes |
| 37 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36448 | -2.956 | -0.5050 | Yes |
| 38 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36458 | -2.975 | -0.4820 | Yes |
| 39 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36495 | -3.045 | -0.4592 | Yes |
| 40 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 36578 | -3.169 | -0.4366 | Yes |
| 41 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36597 | -3.196 | -0.4122 | Yes |
| 42 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36619 | -3.233 | -0.3875 | Yes |
| 43 | DTL | denticleless E3 ubiquitin protein ligase homolog [Source:HGNC Symbol;Acc:HGNC:30288] | 36622 | -3.236 | -0.3623 | Yes |
| 44 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36647 | -3.278 | -0.3373 | Yes |
| 45 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36684 | -3.340 | -0.3122 | Yes |
| 46 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36735 | -3.416 | -0.2869 | Yes |
| 47 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36802 | -3.541 | -0.2610 | Yes |
| 48 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36819 | -3.570 | -0.2336 | Yes |
| 49 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36831 | -3.600 | -0.2058 | Yes |
| 50 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36856 | -3.657 | -0.1779 | Yes |
| 51 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36868 | -3.678 | -0.1495 | Yes |
| 52 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36886 | -3.735 | -0.1207 | Yes |
| 53 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36915 | -3.854 | -0.0914 | Yes |
| 54 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 36946 | -3.945 | -0.0614 | Yes |
| 55 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 37012 | -4.172 | -0.0306 | Yes |
| 56 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 37016 | -4.184 | 0.0020 | Yes |