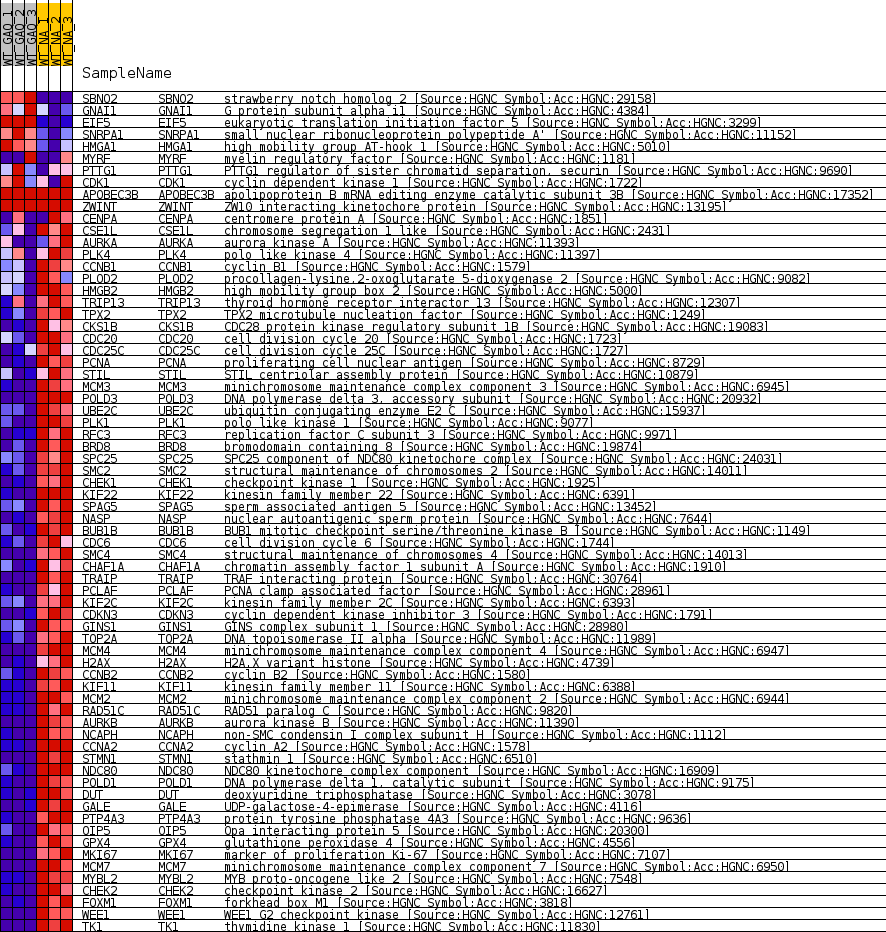
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
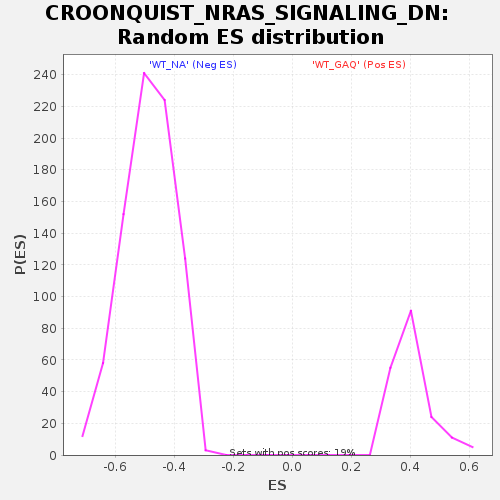
| GeneSet | CROONQUIST_NRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.80617404 |
| Normalized Enrichment Score (NES) | -1.6593438 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.00507687 |
| FWER p-Value | 0.083 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 171 | 3.167 | 0.0225 | No |
| 2 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 2109 | 0.853 | -0.0225 | No |
| 3 | EIF5 | eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] | 2173 | 0.827 | -0.0171 | No |
| 4 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 2230 | 0.810 | -0.0117 | No |
| 5 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 3261 | 0.531 | -0.0350 | No |
| 6 | MYRF | myelin regulatory factor [Source:HGNC Symbol;Acc:HGNC:1181] | 4780 | 0.266 | -0.0737 | No |
| 7 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 5637 | 0.136 | -0.0957 | No |
| 8 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 6135 | 0.073 | -0.1085 | No |
| 9 | APOBEC3B | apolipoprotein B mRNA editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] | 12715 | 0.000 | -0.2862 | No |
| 10 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26168 | 0.000 | -0.6495 | No |
| 11 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 30362 | -0.359 | -0.7597 | No |
| 12 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 30803 | -0.423 | -0.7680 | No |
| 13 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 31738 | -0.568 | -0.7883 | No |
| 14 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 32153 | -0.642 | -0.7940 | No |
| 15 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 32260 | -0.666 | -0.7912 | No |
| 16 | PLOD2 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 [Source:HGNC Symbol;Acc:HGNC:9082] | 32351 | -0.689 | -0.7877 | No |
| 17 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 32479 | -0.717 | -0.7850 | No |
| 18 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33264 | -0.912 | -0.7984 | Yes |
| 19 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 33320 | -0.929 | -0.7919 | Yes |
| 20 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 33639 | -1.014 | -0.7918 | Yes |
| 21 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 33672 | -1.025 | -0.7839 | Yes |
| 22 | CDC25C | cell division cycle 25C [Source:HGNC Symbol;Acc:HGNC:1727] | 33986 | -1.112 | -0.7828 | Yes |
| 23 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 34110 | -1.145 | -0.7763 | Yes |
| 24 | STIL | STIL centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:10879] | 34485 | -1.287 | -0.7754 | Yes |
| 25 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34716 | -1.383 | -0.7698 | Yes |
| 26 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34734 | -1.392 | -0.7583 | Yes |
| 27 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 34791 | -1.414 | -0.7477 | Yes |
| 28 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 34837 | -1.440 | -0.7366 | Yes |
| 29 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 34852 | -1.446 | -0.7246 | Yes |
| 30 | BRD8 | bromodomain containing 8 [Source:HGNC Symbol;Acc:HGNC:19874] | 34946 | -1.490 | -0.7143 | Yes |
| 31 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 35095 | -1.566 | -0.7049 | Yes |
| 32 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35127 | -1.583 | -0.6922 | Yes |
| 33 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 35131 | -1.586 | -0.6787 | Yes |
| 34 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 35249 | -1.649 | -0.6677 | Yes |
| 35 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 35259 | -1.656 | -0.6538 | Yes |
| 36 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35268 | -1.661 | -0.6398 | Yes |
| 37 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35320 | -1.686 | -0.6267 | Yes |
| 38 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35404 | -1.737 | -0.6141 | Yes |
| 39 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35408 | -1.739 | -0.5992 | Yes |
| 40 | CHAF1A | chromatin assembly factor 1 subunit A [Source:HGNC Symbol;Acc:HGNC:1910] | 35426 | -1.748 | -0.5847 | Yes |
| 41 | TRAIP | TRAF interacting protein [Source:HGNC Symbol;Acc:HGNC:30764] | 35458 | -1.764 | -0.5705 | Yes |
| 42 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 35494 | -1.788 | -0.5561 | Yes |
| 43 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 35561 | -1.824 | -0.5422 | Yes |
| 44 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 35605 | -1.855 | -0.5275 | Yes |
| 45 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 35614 | -1.860 | -0.5118 | Yes |
| 46 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35626 | -1.864 | -0.4961 | Yes |
| 47 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 35681 | -1.914 | -0.4812 | Yes |
| 48 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 35698 | -1.925 | -0.4651 | Yes |
| 49 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 35787 | -1.989 | -0.4505 | Yes |
| 50 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35789 | -1.990 | -0.4335 | Yes |
| 51 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36041 | -2.186 | -0.4215 | Yes |
| 52 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 36081 | -2.221 | -0.4035 | Yes |
| 53 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36153 | -2.285 | -0.3859 | Yes |
| 54 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36164 | -2.301 | -0.3664 | Yes |
| 55 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36192 | -2.319 | -0.3473 | Yes |
| 56 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36274 | -2.405 | -0.3289 | Yes |
| 57 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36364 | -2.499 | -0.3099 | Yes |
| 58 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36434 | -2.597 | -0.2895 | Yes |
| 59 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 36479 | -2.649 | -0.2680 | Yes |
| 60 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 36498 | -2.680 | -0.2455 | Yes |
| 61 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36552 | -2.766 | -0.2233 | Yes |
| 62 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 36576 | -2.796 | -0.1999 | Yes |
| 63 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36597 | -2.824 | -0.1763 | Yes |
| 64 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36625 | -2.872 | -0.1524 | Yes |
| 65 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36636 | -2.885 | -0.1280 | Yes |
| 66 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36648 | -2.896 | -0.1035 | Yes |
| 67 | CHEK2 | checkpoint kinase 2 [Source:HGNC Symbol;Acc:HGNC:16627] | 36675 | -2.952 | -0.0789 | Yes |
| 68 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36680 | -2.959 | -0.0536 | Yes |
| 69 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 36855 | -3.309 | -0.0300 | Yes |
| 70 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37054 | -4.241 | 0.0010 | Yes |