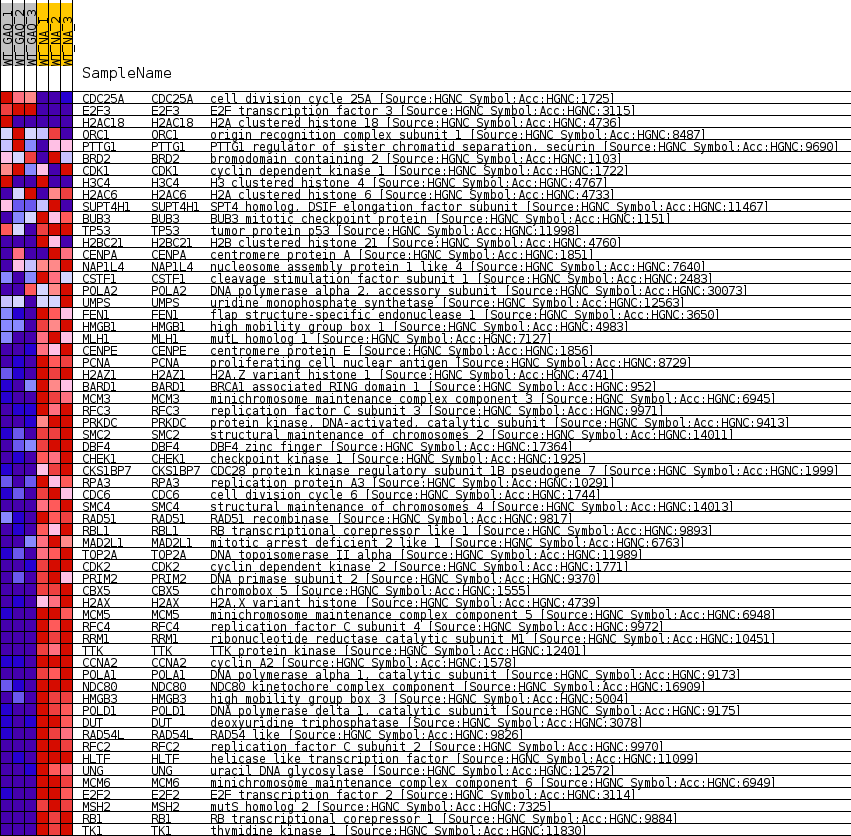
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | REN_BOUND_BY_E2F |
| Enrichment Score (ES) | -0.8280574 |
| Normalized Enrichment Score (NES) | -1.6576059 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.005224461 |
| FWER p-Value | 0.091 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CDC25A | cell division cycle 25A [Source:HGNC Symbol;Acc:HGNC:1725] | 1852 | 0.965 | -0.0401 | No |
| 2 | E2F3 | E2F transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:3115] | 1909 | 0.940 | -0.0319 | No |
| 3 | H2AC18 | H2A clustered histone 18 [Source:HGNC Symbol;Acc:HGNC:4736] | 5245 | 0.194 | -0.1199 | No |
| 4 | ORC1 | origin recognition complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:8487] | 5294 | 0.186 | -0.1193 | No |
| 5 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 5637 | 0.136 | -0.1272 | No |
| 6 | BRD2 | bromodomain containing 2 [Source:HGNC Symbol;Acc:HGNC:1103] | 6063 | 0.081 | -0.1378 | No |
| 7 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 6135 | 0.073 | -0.1390 | No |
| 8 | H3C4 | H3 clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4767] | 6551 | 0.021 | -0.1500 | No |
| 9 | H2AC6 | H2A clustered histone 6 [Source:HGNC Symbol;Acc:HGNC:4733] | 27562 | -0.016 | -0.7172 | No |
| 10 | SUPT4H1 | SPT4 homolog, DSIF elongation factor subunit [Source:HGNC Symbol;Acc:HGNC:11467] | 27761 | -0.040 | -0.7221 | No |
| 11 | BUB3 | BUB3 mitotic checkpoint protein [Source:HGNC Symbol;Acc:HGNC:1151] | 28342 | -0.112 | -0.7366 | No |
| 12 | TP53 | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 29665 | -0.274 | -0.7695 | No |
| 13 | H2BC21 | H2B clustered histone 21 [Source:HGNC Symbol;Acc:HGNC:4760] | 29833 | -0.292 | -0.7710 | No |
| 14 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 30362 | -0.359 | -0.7816 | No |
| 15 | NAP1L4 | nucleosome assembly protein 1 like 4 [Source:HGNC Symbol;Acc:HGNC:7640] | 30549 | -0.385 | -0.7826 | No |
| 16 | CSTF1 | cleavage stimulation factor subunit 1 [Source:HGNC Symbol;Acc:HGNC:2483] | 31205 | -0.485 | -0.7953 | No |
| 17 | POLA2 | DNA polymerase alpha 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:30073] | 31413 | -0.515 | -0.7956 | No |
| 18 | UMPS | uridine monophosphate synthetase [Source:HGNC Symbol;Acc:HGNC:12563] | 32264 | -0.667 | -0.8117 | No |
| 19 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 32680 | -0.764 | -0.8150 | No |
| 20 | HMGB1 | high mobility group box 1 [Source:HGNC Symbol;Acc:HGNC:4983] | 33164 | -0.884 | -0.8189 | Yes |
| 21 | MLH1 | mutL homolog 1 [Source:HGNC Symbol;Acc:HGNC:7127] | 33190 | -0.890 | -0.8104 | Yes |
| 22 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 33831 | -1.063 | -0.8168 | Yes |
| 23 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 34110 | -1.145 | -0.8125 | Yes |
| 24 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34211 | -1.183 | -0.8030 | Yes |
| 25 | BARD1 | BRCA1 associated RING domain 1 [Source:HGNC Symbol;Acc:HGNC:952] | 34661 | -1.354 | -0.8011 | Yes |
| 26 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34716 | -1.383 | -0.7883 | Yes |
| 27 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 34852 | -1.446 | -0.7771 | Yes |
| 28 | PRKDC | protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413] | 35043 | -1.541 | -0.7663 | Yes |
| 29 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35127 | -1.583 | -0.7523 | Yes |
| 30 | DBF4 | DBF4 zinc finger [Source:HGNC Symbol;Acc:HGNC:17364] | 35128 | -1.584 | -0.7359 | Yes |
| 31 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 35131 | -1.586 | -0.7196 | Yes |
| 32 | CKS1BP7 | CDC28 protein kinase regulatory subunit 1B pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:1999] | 35152 | -1.601 | -0.7037 | Yes |
| 33 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 35321 | -1.686 | -0.6908 | Yes |
| 34 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35404 | -1.737 | -0.6751 | Yes |
| 35 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35408 | -1.739 | -0.6573 | Yes |
| 36 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 35435 | -1.752 | -0.6400 | Yes |
| 37 | RBL1 | RB transcriptional corepressor like 1 [Source:HGNC Symbol;Acc:HGNC:9893] | 35503 | -1.793 | -0.6233 | Yes |
| 38 | MAD2L1 | mitotic arrest deficient 2 like 1 [Source:HGNC Symbol;Acc:HGNC:6763] | 35525 | -1.805 | -0.6052 | Yes |
| 39 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35626 | -1.864 | -0.5887 | Yes |
| 40 | CDK2 | cyclin dependent kinase 2 [Source:HGNC Symbol;Acc:HGNC:1771] | 35643 | -1.882 | -0.5698 | Yes |
| 41 | PRIM2 | DNA primase subunit 2 [Source:HGNC Symbol;Acc:HGNC:9370] | 35686 | -1.917 | -0.5511 | Yes |
| 42 | CBX5 | chromobox 5 [Source:HGNC Symbol;Acc:HGNC:1555] | 35694 | -1.922 | -0.5315 | Yes |
| 43 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 35698 | -1.925 | -0.5118 | Yes |
| 44 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35722 | -1.939 | -0.4924 | Yes |
| 45 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35922 | -2.100 | -0.4761 | Yes |
| 46 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35995 | -2.149 | -0.4559 | Yes |
| 47 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36013 | -2.163 | -0.4341 | Yes |
| 48 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36192 | -2.319 | -0.4150 | Yes |
| 49 | POLA1 | DNA polymerase alpha 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9173] | 36270 | -2.402 | -0.3923 | Yes |
| 50 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36364 | -2.499 | -0.3691 | Yes |
| 51 | HMGB3 | high mobility group box 3 [Source:HGNC Symbol;Acc:HGNC:5004] | 36397 | -2.543 | -0.3437 | Yes |
| 52 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36434 | -2.597 | -0.3179 | Yes |
| 53 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 36479 | -2.649 | -0.2918 | Yes |
| 54 | RAD54L | RAD54 like [Source:HGNC Symbol;Acc:HGNC:9826] | 36522 | -2.718 | -0.2649 | Yes |
| 55 | RFC2 | replication factor C subunit 2 [Source:HGNC Symbol;Acc:HGNC:9970] | 36539 | -2.738 | -0.2371 | Yes |
| 56 | HLTF | helicase like transcription factor [Source:HGNC Symbol;Acc:HGNC:11099] | 36562 | -2.773 | -0.2092 | Yes |
| 57 | UNG | uracil DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:12572] | 36823 | -3.229 | -0.1829 | Yes |
| 58 | MCM6 | minichromosome maintenance complex component 6 [Source:HGNC Symbol;Acc:HGNC:6949] | 36860 | -3.318 | -0.1497 | Yes |
| 59 | E2F2 | E2F transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:3114] | 36862 | -3.322 | -0.1155 | Yes |
| 60 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 36967 | -3.703 | -0.0801 | Yes |
| 61 | RB1 | RB transcriptional corepressor 1 [Source:HGNC Symbol;Acc:HGNC:9884] | 36995 | -3.849 | -0.0412 | Yes |
| 62 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37054 | -4.241 | 0.0010 | Yes |