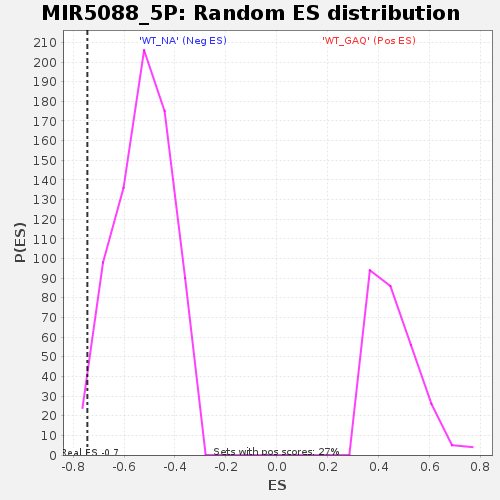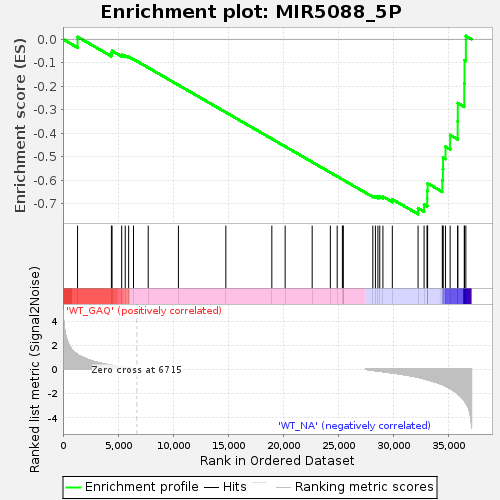
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | MIR5088_5P |
| Enrichment Score (ES) | -0.7443125 |
| Normalized Enrichment Score (NES) | -1.4138551 |
| Nominal p-value | 0.013717421 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | USP14 | ubiquitin specific peptidase 14 [Source:HGNC Symbol;Acc:HGNC:12612] | 1321 | 1.198 | 0.0101 | No |
| 2 | AKAP3 | A-kinase anchoring protein 3 [Source:HGNC Symbol;Acc:HGNC:373] | 4394 | 0.324 | -0.0604 | No |
| 3 | HECW2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:29853] | 4452 | 0.316 | -0.0499 | No |
| 4 | CTDNEP1 | CTD nuclear envelope phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:19085] | 5326 | 0.181 | -0.0665 | No |
| 5 | MSANTD2 | Myb/SANT DNA binding domain containing 2 [Source:HGNC Symbol;Acc:HGNC:26266] | 5650 | 0.135 | -0.0701 | No |
| 6 | ZNF135 | zinc finger protein 135 [Source:HGNC Symbol;Acc:HGNC:12919] | 5960 | 0.092 | -0.0749 | No |
| 7 | ATPAF1 | ATP synthase mitochondrial F1 complex assembly factor 1 [Source:HGNC Symbol;Acc:HGNC:18803] | 6400 | 0.039 | -0.0852 | No |
| 8 | CNTNAP2 | contactin associated protein 2 [Source:HGNC Symbol;Acc:HGNC:13830] | 7736 | 0.000 | -0.1213 | No |
| 9 | SLC22A2 | solute carrier family 22 member 2 [Source:HGNC Symbol;Acc:HGNC:10966] | 10477 | 0.000 | -0.1952 | No |
| 10 | PCDH10 | protocadherin 10 [Source:HGNC Symbol;Acc:HGNC:13404] | 14782 | 0.000 | -0.3114 | No |
| 11 | MIP | major intrinsic protein of lens fiber [Source:HGNC Symbol;Acc:HGNC:7103] | 18958 | 0.000 | -0.4241 | No |
| 12 | TMEM47 | transmembrane protein 47 [Source:HGNC Symbol;Acc:HGNC:18515] | 20167 | 0.000 | -0.4567 | No |
| 13 | PI15 | peptidase inhibitor 15 [Source:HGNC Symbol;Acc:HGNC:8946] | 22620 | 0.000 | -0.5228 | No |
| 14 | ADCYAP1R1 | ADCYAP receptor type I [Source:HGNC Symbol;Acc:HGNC:242] | 24271 | 0.000 | -0.5674 | No |
| 15 | KCNMB2 | potassium calcium-activated channel subfamily M regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6286] | 24890 | 0.000 | -0.5840 | No |
| 16 | PRSS8 | serine protease 8 [Source:HGNC Symbol;Acc:HGNC:9491] | 25362 | 0.000 | -0.5968 | No |
| 17 | RIMS4 | regulating synaptic membrane exocytosis 4 [Source:HGNC Symbol;Acc:HGNC:16183] | 25425 | 0.000 | -0.5984 | No |
| 18 | FGF9 | fibroblast growth factor 9 [Source:HGNC Symbol;Acc:HGNC:3687] | 25429 | 0.000 | -0.5985 | No |
| 19 | CANX | calnexin [Source:HGNC Symbol;Acc:HGNC:1473] | 28124 | -0.084 | -0.6680 | No |
| 20 | MECP2 | methyl-CpG binding protein 2 [Source:HGNC Symbol;Acc:HGNC:6990] | 28360 | -0.114 | -0.6700 | No |
| 21 | KDM2A | lysine demethylase 2A [Source:HGNC Symbol;Acc:HGNC:13606] | 28587 | -0.138 | -0.6708 | No |
| 22 | HIC2 | HIC ZBTB transcriptional repressor 2 [Source:HGNC Symbol;Acc:HGNC:18595] | 28756 | -0.157 | -0.6694 | No |
| 23 | LRRTM2 | leucine rich repeat transmembrane neuronal 2 [Source:HGNC Symbol;Acc:HGNC:19409] | 29042 | -0.194 | -0.6697 | No |
| 24 | PSMB9 | proteasome 20S subunit beta 9 [Source:HGNC Symbol;Acc:HGNC:9546] | 29896 | -0.298 | -0.6813 | No |
| 25 | PJA1 | praja ring finger ubiquitin ligase 1 [Source:HGNC Symbol;Acc:HGNC:16648] | 32232 | -0.662 | -0.7190 | Yes |
| 26 | BZW2 | basic leucine zipper and W2 domains 2 [Source:HGNC Symbol;Acc:HGNC:18808] | 32779 | -0.787 | -0.7037 | Yes |
| 27 | PTBP2 | polypyrimidine tract binding protein 2 [Source:HGNC Symbol;Acc:HGNC:17662] | 33063 | -0.858 | -0.6785 | Yes |
| 28 | GALNT2 | polypeptide N-acetylgalactosaminyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:4124] | 33065 | -0.859 | -0.6457 | Yes |
| 29 | VAPB | VAMP associated protein B and C [Source:HGNC Symbol;Acc:HGNC:12649] | 33101 | -0.867 | -0.6136 | Yes |
| 30 | PTER | phosphotriesterase related [Source:HGNC Symbol;Acc:HGNC:9590] | 34433 | -1.265 | -0.6012 | Yes |
| 31 | IDS | iduronate 2-sulfatase [Source:HGNC Symbol;Acc:HGNC:5389] | 34486 | -1.288 | -0.5534 | Yes |
| 32 | XRCC6 | X-ray repair cross complementing 6 [Source:HGNC Symbol;Acc:HGNC:4055] | 34501 | -1.292 | -0.5044 | Yes |
| 33 | THTPA | thiamine triphosphatase [Source:HGNC Symbol;Acc:HGNC:18987] | 34720 | -1.384 | -0.4573 | Yes |
| 34 | FOXO4 | forkhead box O4 [Source:HGNC Symbol;Acc:HGNC:7139] | 35144 | -1.596 | -0.4078 | Yes |
| 35 | CENPI | centromere protein I [Source:HGNC Symbol;Acc:HGNC:3968] | 35829 | -2.021 | -0.3491 | Yes |
| 36 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 [Source:HGNC Symbol;Acc:HGNC:16922] | 35839 | -2.028 | -0.2718 | Yes |
| 37 | SH3PXD2A | SH3 and PX domains 2A [Source:HGNC Symbol;Acc:HGNC:23664] | 36427 | -2.586 | -0.1888 | Yes |
| 38 | FUT10 | fucosyltransferase 10 [Source:HGNC Symbol;Acc:HGNC:19234] | 36451 | -2.619 | -0.0894 | Yes |
| 39 | PMP22 | peripheral myelin protein 22 [Source:HGNC Symbol;Acc:HGNC:9118] | 36571 | -2.790 | 0.0140 | Yes |