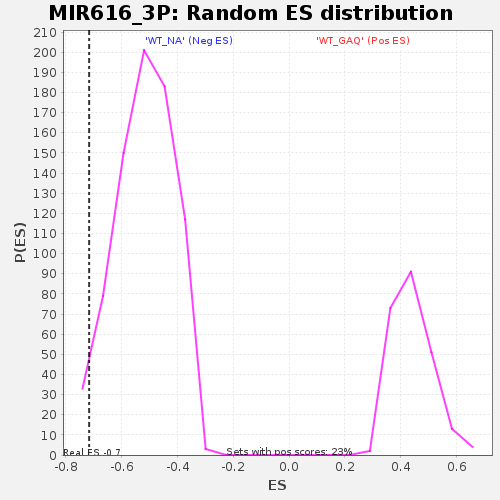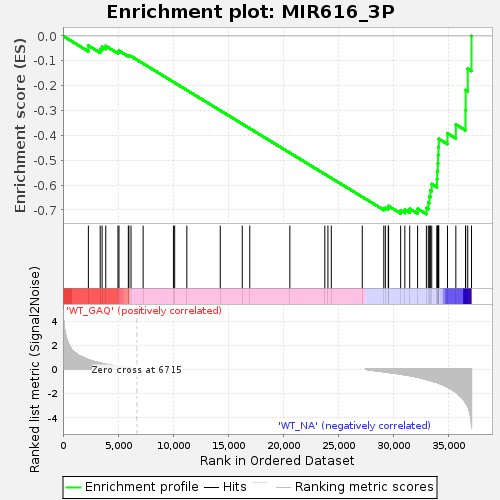
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | MIR616_3P |
| Enrichment Score (ES) | -0.7170002 |
| Normalized Enrichment Score (NES) | -1.3829012 |
| Nominal p-value | 0.028720627 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SLC17A8 | solute carrier family 17 member 8 [Source:HGNC Symbol;Acc:HGNC:20151] | 2304 | 0.785 | -0.0390 | No |
| 2 | CCDC141 | coiled-coil domain containing 141 [Source:HGNC Symbol;Acc:HGNC:26821] | 3375 | 0.505 | -0.0529 | No |
| 3 | SERPINH1 | serpin family H member 1 [Source:HGNC Symbol;Acc:HGNC:1546] | 3548 | 0.470 | -0.0437 | No |
| 4 | NCL | nucleolin [Source:HGNC Symbol;Acc:HGNC:7667] | 3882 | 0.410 | -0.0406 | No |
| 5 | TXK | TXK tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:12434] | 4990 | 0.233 | -0.0636 | No |
| 6 | ARL8A | ADP ribosylation factor like GTPase 8A [Source:HGNC Symbol;Acc:HGNC:25192] | 5082 | 0.219 | -0.0596 | No |
| 7 | LMAN1 | lectin, mannose binding 1 [Source:HGNC Symbol;Acc:HGNC:6631] | 5930 | 0.095 | -0.0796 | No |
| 8 | PRKAG1 | protein kinase AMP-activated non-catalytic subunit gamma 1 [Source:HGNC Symbol;Acc:HGNC:9385] | 5981 | 0.089 | -0.0783 | No |
| 9 | BIRC3 | baculoviral IAP repeat containing 3 [Source:HGNC Symbol;Acc:HGNC:591] | 6174 | 0.068 | -0.0815 | No |
| 10 | FGF14 | fibroblast growth factor 14 [Source:HGNC Symbol;Acc:HGNC:3671] | 7273 | 0.000 | -0.1111 | No |
| 11 | KCNA4 | potassium voltage-gated channel subfamily A member 4 [Source:HGNC Symbol;Acc:HGNC:6222] | 10024 | 0.000 | -0.1854 | No |
| 12 | KCNJ1 | potassium inwardly rectifying channel subfamily J member 1 [Source:HGNC Symbol;Acc:HGNC:6255] | 10137 | 0.000 | -0.1884 | No |
| 13 | RAB27B | RAB27B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9767] | 11242 | 0.000 | -0.2182 | No |
| 14 | PIK3C2B | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta [Source:HGNC Symbol;Acc:HGNC:8972] | 14276 | 0.000 | -0.3001 | No |
| 15 | GC | GC vitamin D binding protein [Source:HGNC Symbol;Acc:HGNC:4187] | 16273 | 0.000 | -0.3540 | No |
| 16 | SYN3 | synapsin III [Source:HGNC Symbol;Acc:HGNC:11496] | 16955 | 0.000 | -0.3723 | No |
| 17 | C4orf19 | chromosome 4 open reading frame 19 [Source:HGNC Symbol;Acc:HGNC:25618] | 20593 | 0.000 | -0.4705 | No |
| 18 | RBM43 | RNA binding motif protein 43 [Source:HGNC Symbol;Acc:HGNC:24790] | 23767 | 0.000 | -0.5562 | No |
| 19 | SCGB1D2 | secretoglobin family 1D member 2 [Source:HGNC Symbol;Acc:HGNC:18396] | 24049 | 0.000 | -0.5638 | No |
| 20 | ADGRG6 | adhesion G protein-coupled receptor G6 [Source:HGNC Symbol;Acc:HGNC:13841] | 24356 | 0.000 | -0.5720 | No |
| 21 | MAEL | maelstrom spermatogenic transposon silencer [Source:HGNC Symbol;Acc:HGNC:25929] | 27168 | 0.000 | -0.6479 | No |
| 22 | ATP2B3 | ATPase plasma membrane Ca2+ transporting 3 [Source:HGNC Symbol;Acc:HGNC:816] | 29121 | -0.204 | -0.6946 | No |
| 23 | MBNL3 | muscleblind like splicing regulator 3 [Source:HGNC Symbol;Acc:HGNC:20564] | 29262 | -0.219 | -0.6919 | No |
| 24 | MAPK9 | mitogen-activated protein kinase 9 [Source:HGNC Symbol;Acc:HGNC:6886] | 29525 | -0.256 | -0.6914 | No |
| 25 | PDE6H | phosphodiesterase 6H [Source:HGNC Symbol;Acc:HGNC:8790] | 29557 | -0.260 | -0.6846 | No |
| 26 | GPBP1L1 | GC-rich promoter binding protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:28843] | 30651 | -0.399 | -0.7023 | Yes |
| 27 | RDH11 | retinol dehydrogenase 11 [Source:HGNC Symbol;Acc:HGNC:17964] | 31032 | -0.458 | -0.6990 | Yes |
| 28 | ERBIN | erbb2 interacting protein [Source:HGNC Symbol;Acc:HGNC:15842] | 31475 | -0.525 | -0.6954 | Yes |
| 29 | MCPH1 | microcephalin 1 [Source:HGNC Symbol;Acc:HGNC:6954] | 32191 | -0.650 | -0.6955 | Yes |
| 30 | ZNF22 | zinc finger protein 22 [Source:HGNC Symbol;Acc:HGNC:13012] | 32988 | -0.839 | -0.6922 | Yes |
| 31 | CDC7 | cell division cycle 7 [Source:HGNC Symbol;Acc:HGNC:1745] | 33160 | -0.882 | -0.6708 | Yes |
| 32 | NR3C1 | nuclear receptor subfamily 3 group C member 1 [Source:HGNC Symbol;Acc:HGNC:7978] | 33260 | -0.911 | -0.6466 | Yes |
| 33 | BRD1 | bromodomain containing 1 [Source:HGNC Symbol;Acc:HGNC:1102] | 33351 | -0.936 | -0.6213 | Yes |
| 34 | ANTXR1 | ANTXR cell adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:21014] | 33474 | -0.966 | -0.5961 | Yes |
| 35 | LONRF3 | LON peptidase N-terminal domain and ring finger 3 [Source:HGNC Symbol;Acc:HGNC:21152] | 33945 | -1.095 | -0.5764 | Yes |
| 36 | TNFSF14 | TNF superfamily member 14 [Source:HGNC Symbol;Acc:HGNC:11930] | 33970 | -1.104 | -0.5445 | Yes |
| 37 | ADD1 | adducin 1 [Source:HGNC Symbol;Acc:HGNC:243] | 34024 | -1.122 | -0.5127 | Yes |
| 38 | DDX59 | DEAD-box helicase 59 [Source:HGNC Symbol;Acc:HGNC:25360] | 34050 | -1.131 | -0.4800 | Yes |
| 39 | NEDD4L | NEDD4 like E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:HGNC:7728] | 34080 | -1.139 | -0.4472 | Yes |
| 40 | DAPK1 | death associated protein kinase 1 [Source:HGNC Symbol;Acc:HGNC:2674] | 34115 | -1.146 | -0.4142 | Yes |
| 41 | NECTIN3 | nectin cell adhesion molecule 3 [Source:HGNC Symbol;Acc:HGNC:17664] | 34901 | -1.471 | -0.3919 | Yes |
| 42 | DCUN1D4 | defective in cullin neddylation 1 domain containing 4 [Source:HGNC Symbol;Acc:HGNC:28998] | 35661 | -1.895 | -0.3564 | Yes |
| 43 | CPED1 | cadherin like and PC-esterase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:26159] | 36540 | -2.739 | -0.2992 | Yes |
| 44 | LSM11 | LSM11, U7 small nuclear RNA associated [Source:HGNC Symbol;Acc:HGNC:30860] | 36556 | -2.768 | -0.2179 | Yes |
| 45 | HPGD | 15-hydroxyprostaglandin dehydrogenase [Source:HGNC Symbol;Acc:HGNC:5154] | 36743 | -3.080 | -0.1319 | Yes |
| 46 | RNF144B | ring finger protein 144B [Source:HGNC Symbol;Acc:HGNC:21578] | 37085 | -4.781 | 0.0001 | Yes |