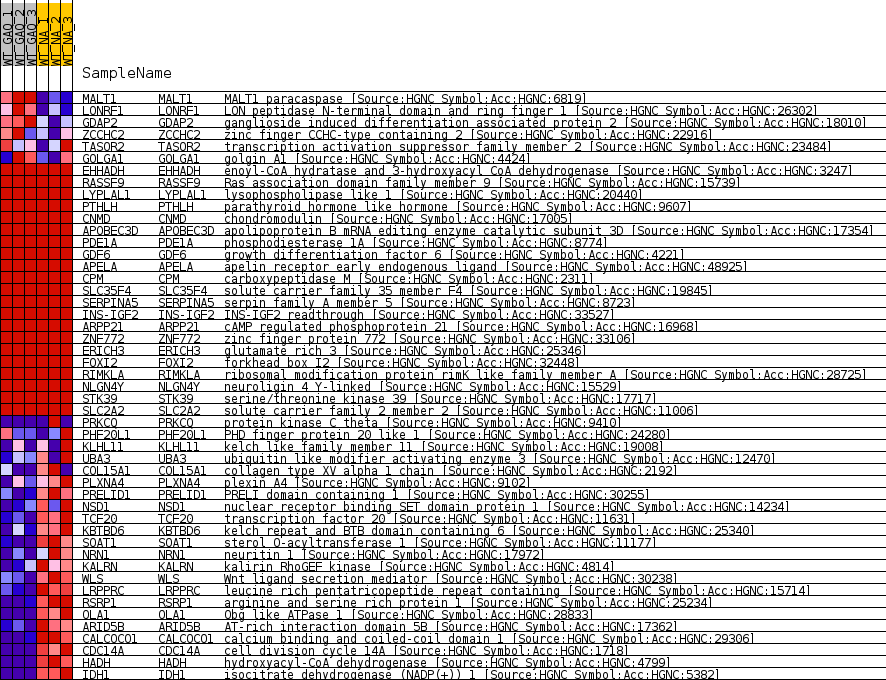
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | MIR6790_5P |
| Enrichment Score (ES) | -0.71976364 |
| Normalized Enrichment Score (NES) | -1.4090474 |
| Nominal p-value | 0.022397893 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MALT1 | MALT1 paracaspase [Source:HGNC Symbol;Acc:HGNC:6819] | 327 | 2.528 | 0.0652 | No |
| 2 | LONRF1 | LON peptidase N-terminal domain and ring finger 1 [Source:HGNC Symbol;Acc:HGNC:26302] | 1225 | 1.270 | 0.0782 | No |
| 3 | GDAP2 | ganglioside induced differentiation associated protein 2 [Source:HGNC Symbol;Acc:HGNC:18010] | 4011 | 0.387 | 0.0144 | No |
| 4 | ZCCHC2 | zinc finger CCHC-type containing 2 [Source:HGNC Symbol;Acc:HGNC:22916] | 4629 | 0.291 | 0.0062 | No |
| 5 | TASOR2 | transcription activation suppressor family member 2 [Source:HGNC Symbol;Acc:HGNC:23484] | 5339 | 0.180 | -0.0077 | No |
| 6 | GOLGA1 | golgin A1 [Source:HGNC Symbol;Acc:HGNC:4424] | 5531 | 0.150 | -0.0084 | No |
| 7 | EHHADH | enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3247] | 8300 | 0.000 | -0.0832 | No |
| 8 | RASSF9 | Ras association domain family member 9 [Source:HGNC Symbol;Acc:HGNC:15739] | 9000 | 0.000 | -0.1020 | No |
| 9 | LYPLAL1 | lysophospholipase like 1 [Source:HGNC Symbol;Acc:HGNC:20440] | 11665 | 0.000 | -0.1739 | No |
| 10 | PTHLH | parathyroid hormone like hormone [Source:HGNC Symbol;Acc:HGNC:9607] | 11895 | 0.000 | -0.1801 | No |
| 11 | CNMD | chondromodulin [Source:HGNC Symbol;Acc:HGNC:17005] | 12522 | 0.000 | -0.1970 | No |
| 12 | APOBEC3D | apolipoprotein B mRNA editing enzyme catalytic subunit 3D [Source:HGNC Symbol;Acc:HGNC:17354] | 12697 | 0.000 | -0.2017 | No |
| 13 | PDE1A | phosphodiesterase 1A [Source:HGNC Symbol;Acc:HGNC:8774] | 13608 | 0.000 | -0.2263 | No |
| 14 | GDF6 | growth differentiation factor 6 [Source:HGNC Symbol;Acc:HGNC:4221] | 13626 | 0.000 | -0.2268 | No |
| 15 | APELA | apelin receptor early endogenous ligand [Source:HGNC Symbol;Acc:HGNC:48925] | 15514 | 0.000 | -0.2777 | No |
| 16 | CPM | carboxypeptidase M [Source:HGNC Symbol;Acc:HGNC:2311] | 15960 | 0.000 | -0.2897 | No |
| 17 | SLC35F4 | solute carrier family 35 member F4 [Source:HGNC Symbol;Acc:HGNC:19845] | 16023 | 0.000 | -0.2914 | No |
| 18 | SERPINA5 | serpin family A member 5 [Source:HGNC Symbol;Acc:HGNC:8723] | 17819 | 0.000 | -0.3398 | No |
| 19 | INS-IGF2 | INS-IGF2 readthrough [Source:HGNC Symbol;Acc:HGNC:33527] | 19975 | 0.000 | -0.3980 | No |
| 20 | ARPP21 | cAMP regulated phosphoprotein 21 [Source:HGNC Symbol;Acc:HGNC:16968] | 20924 | 0.000 | -0.4236 | No |
| 21 | ZNF772 | zinc finger protein 772 [Source:HGNC Symbol;Acc:HGNC:33106] | 20986 | 0.000 | -0.4253 | No |
| 22 | ERICH3 | glutamate rich 3 [Source:HGNC Symbol;Acc:HGNC:25346] | 21268 | 0.000 | -0.4328 | No |
| 23 | FOXI2 | forkhead box I2 [Source:HGNC Symbol;Acc:HGNC:32448] | 23626 | 0.000 | -0.4965 | No |
| 24 | RIMKLA | ribosomal modification protein rimK like family member A [Source:HGNC Symbol;Acc:HGNC:28725] | 23648 | 0.000 | -0.4970 | No |
| 25 | NLGN4Y | neuroligin 4 Y-linked [Source:HGNC Symbol;Acc:HGNC:15529] | 25506 | 0.000 | -0.5472 | No |
| 26 | STK39 | serine/threonine kinase 39 [Source:HGNC Symbol;Acc:HGNC:17717] | 25823 | 0.000 | -0.5557 | No |
| 27 | SLC2A2 | solute carrier family 2 member 2 [Source:HGNC Symbol;Acc:HGNC:11006] | 27450 | 0.000 | -0.5996 | No |
| 28 | PRKCQ | protein kinase C theta [Source:HGNC Symbol;Acc:HGNC:9410] | 27925 | -0.063 | -0.6106 | No |
| 29 | PHF20L1 | PHD finger protein 20 like 1 [Source:HGNC Symbol;Acc:HGNC:24280] | 27990 | -0.067 | -0.6103 | No |
| 30 | KLHL11 | kelch like family member 11 [Source:HGNC Symbol;Acc:HGNC:19008] | 28300 | -0.108 | -0.6155 | No |
| 31 | UBA3 | ubiquitin like modifier activating enzyme 3 [Source:HGNC Symbol;Acc:HGNC:12470] | 29501 | -0.252 | -0.6405 | No |
| 32 | COL15A1 | collagen type XV alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:2192] | 31812 | -0.578 | -0.6860 | No |
| 33 | PLXNA4 | plexin A4 [Source:HGNC Symbol;Acc:HGNC:9102] | 33064 | -0.859 | -0.6946 | Yes |
| 34 | PRELID1 | PRELI domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30255] | 33233 | -0.903 | -0.6727 | Yes |
| 35 | NSD1 | nuclear receptor binding SET domain protein 1 [Source:HGNC Symbol;Acc:HGNC:14234] | 33325 | -0.930 | -0.6479 | Yes |
| 36 | TCF20 | transcription factor 20 [Source:HGNC Symbol;Acc:HGNC:11631] | 33442 | -0.958 | -0.6230 | Yes |
| 37 | KBTBD6 | kelch repeat and BTB domain containing 6 [Source:HGNC Symbol;Acc:HGNC:25340] | 33585 | -0.997 | -0.5976 | Yes |
| 38 | SOAT1 | sterol O-acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:11177] | 33748 | -1.043 | -0.5715 | Yes |
| 39 | NRN1 | neuritin 1 [Source:HGNC Symbol;Acc:HGNC:17972] | 33798 | -1.053 | -0.5419 | Yes |
| 40 | KALRN | kalirin RhoGEF kinase [Source:HGNC Symbol;Acc:HGNC:4814] | 34042 | -1.128 | -0.5155 | Yes |
| 41 | WLS | Wnt ligand secretion mediator [Source:HGNC Symbol;Acc:HGNC:30238] | 34081 | -1.140 | -0.4831 | Yes |
| 42 | LRPPRC | leucine rich pentatricopeptide repeat containing [Source:HGNC Symbol;Acc:HGNC:15714] | 34617 | -1.338 | -0.4584 | Yes |
| 43 | RSRP1 | arginine and serine rich protein 1 [Source:HGNC Symbol;Acc:HGNC:25234] | 35098 | -1.567 | -0.4254 | Yes |
| 44 | OLA1 | Obg like ATPase 1 [Source:HGNC Symbol;Acc:HGNC:28833] | 35292 | -1.671 | -0.3817 | Yes |
| 45 | ARID5B | AT-rich interaction domain 5B [Source:HGNC Symbol;Acc:HGNC:17362] | 35487 | -1.784 | -0.3347 | Yes |
| 46 | CALCOCO1 | calcium binding and coiled-coil domain 1 [Source:HGNC Symbol;Acc:HGNC:29306] | 36422 | -2.575 | -0.2845 | Yes |
| 47 | CDC14A | cell division cycle 14A [Source:HGNC Symbol;Acc:HGNC:1718] | 36687 | -2.974 | -0.2045 | Yes |
| 48 | HADH | hydroxyacyl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4799] | 36927 | -3.577 | -0.1062 | Yes |
| 49 | IDH1 | isocitrate dehydrogenase (NADP(+)) 1 [Source:HGNC Symbol;Acc:HGNC:5382] | 36979 | -3.774 | 0.0030 | Yes |