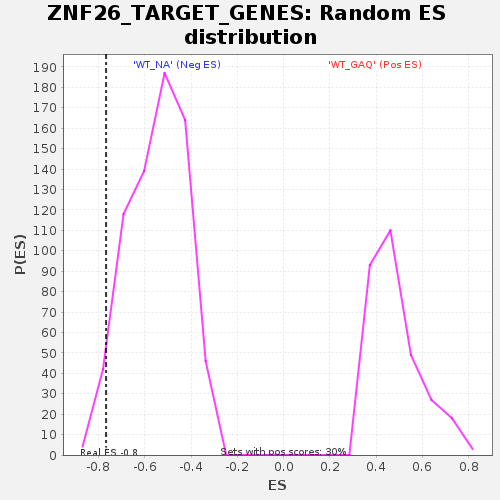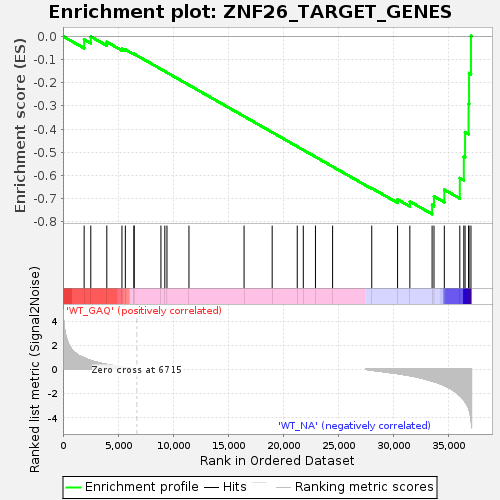
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | ZNF26_TARGET_GENES |
| Enrichment Score (ES) | -0.7669024 |
| Normalized Enrichment Score (NES) | -1.4009058 |
| Nominal p-value | 0.03142857 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | COX16 | cytochrome c oxidase assembly factor COX16 [Source:HGNC Symbol;Acc:HGNC:20213] | 1919 | 0.930 | -0.0141 | No |
| 2 | CLCN5 | chloride voltage-gated channel 5 [Source:HGNC Symbol;Acc:HGNC:2023] | 2522 | 0.718 | -0.0013 | No |
| 3 | TAGLN2P1 | transgelin 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21739] | 3977 | 0.393 | -0.0246 | No |
| 4 | STAT6 | signal transducer and activator of transcription 6 [Source:HGNC Symbol;Acc:HGNC:11368] | 5352 | 0.177 | -0.0545 | No |
| 5 | LYPLA2P3 | LYPLA2 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:50446] | 5659 | 0.133 | -0.0574 | No |
| 6 | SNX16 | sorting nexin 16 [Source:HGNC Symbol;Acc:HGNC:14980] | 6440 | 0.034 | -0.0771 | No |
| 7 | OTOS | otospiralin [Source:HGNC Symbol;Acc:HGNC:22644] | 6459 | 0.032 | -0.0763 | No |
| 8 | LINC01251 | long intergenic non-protein coding RNA 1251 [Source:HGNC Symbol;Acc:HGNC:49845] | 8889 | 0.000 | -0.1418 | No |
| 9 | LINC01119 | long intergenic non-protein coding RNA 1119 [Source:HGNC Symbol;Acc:HGNC:49262] | 9244 | 0.000 | -0.1513 | No |
| 10 | SATB1-AS1 | SATB1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:50687] | 9438 | 0.000 | -0.1566 | No |
| 11 | SRGAP1 | SLIT-ROBO Rho GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:17382] | 11433 | 0.000 | -0.2104 | No |
| 12 | ZNF561-AS1 | ZNF561 antisense RNA 1 (head to head) [Source:HGNC Symbol;Acc:HGNC:27613] | 16437 | 0.000 | -0.3453 | No |
| 13 | OSMR | oncostatin M receptor [Source:HGNC Symbol;Acc:HGNC:8507] | 18989 | 0.000 | -0.4142 | No |
| 14 | ERICH1 | glutamate rich 1 [Source:HGNC Symbol;Acc:HGNC:27234] | 21265 | 0.000 | -0.4756 | No |
| 15 | ZNF567 | zinc finger protein 567 [Source:HGNC Symbol;Acc:HGNC:28696] | 21813 | 0.000 | -0.4903 | No |
| 16 | CCL8 | C-C motif chemokine ligand 8 [Source:HGNC Symbol;Acc:HGNC:10635] | 22918 | 0.000 | -0.5201 | No |
| 17 | LINC00431 | long intergenic non-protein coding RNA 431 [Source:HGNC Symbol;Acc:HGNC:42766] | 24475 | 0.000 | -0.5621 | No |
| 18 | FABP5P7 | fatty acid binding protein 5 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:31070] | 28027 | -0.070 | -0.6551 | No |
| 19 | WWP2 | WW domain containing E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:16804] | 30371 | -0.360 | -0.7037 | No |
| 20 | GSTO1 | glutathione S-transferase omega 1 [Source:HGNC Symbol;Acc:HGNC:13312] | 31487 | -0.528 | -0.7125 | No |
| 21 | OSBPL9 | oxysterol binding protein like 9 [Source:HGNC Symbol;Acc:HGNC:16386] | 33506 | -0.976 | -0.7274 | Yes |
| 22 | ZNF45 | zinc finger protein 45 [Source:HGNC Symbol;Acc:HGNC:13111] | 33679 | -1.028 | -0.6905 | Yes |
| 23 | UAP1 | UDP-N-acetylglucosamine pyrophosphorylase 1 [Source:HGNC Symbol;Acc:HGNC:12457] | 34616 | -1.338 | -0.6616 | Yes |
| 24 | UHRF1 | ubiquitin like with PHD and ring finger domains 1 [Source:HGNC Symbol;Acc:HGNC:12556] | 36022 | -2.169 | -0.6117 | Yes |
| 25 | BBC3 | BCL2 binding component 3 [Source:HGNC Symbol;Acc:HGNC:17868] | 36385 | -2.525 | -0.5192 | Yes |
| 26 | MPPE1 | metallophosphoesterase 1 [Source:HGNC Symbol;Acc:HGNC:15988] | 36500 | -2.681 | -0.4138 | Yes |
| 27 | OMG | oligodendrocyte myelin glycoprotein [Source:HGNC Symbol;Acc:HGNC:8135] | 36817 | -3.215 | -0.2922 | Yes |
| 28 | E2F2 | E2F transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:3114] | 36862 | -3.322 | -0.1589 | Yes |
| 29 | PFKM | phosphofructokinase, muscle [Source:HGNC Symbol;Acc:HGNC:8877] | 37035 | -4.078 | 0.0015 | Yes |