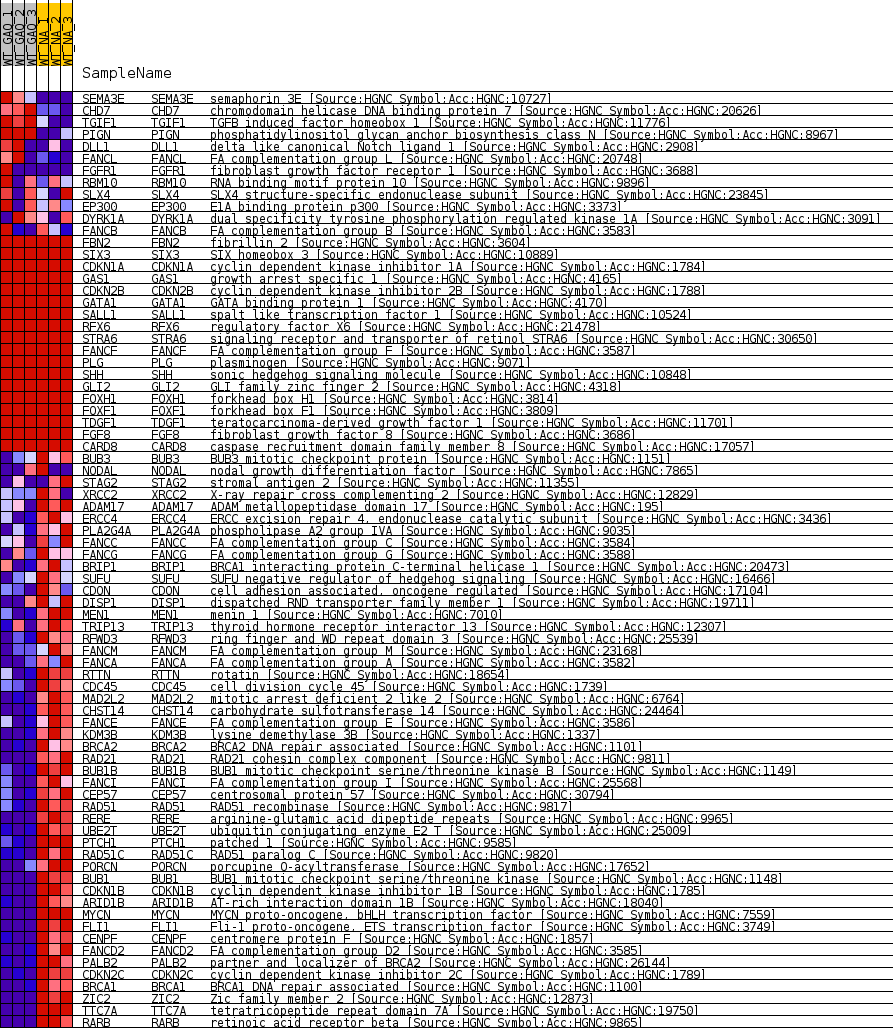
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_GAQ_versus_WT_NA.greenbergcls.cls #WT_GAQ_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_GAQ_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | HP_ABNORMAL_DUODENUM_MORPHOLOGY |
| Enrichment Score (ES) | -0.7734935 |
| Normalized Enrichment Score (NES) | -1.6098392 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06742532 |
| FWER p-Value | 0.584 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SEMA3E | semaphorin 3E [Source:HGNC Symbol;Acc:HGNC:10727] | 1240 | 1.257 | -0.0185 | No |
| 2 | CHD7 | chromodomain helicase DNA binding protein 7 [Source:HGNC Symbol;Acc:HGNC:20626] | 1374 | 1.165 | -0.0081 | No |
| 3 | TGIF1 | TGFB induced factor homeobox 1 [Source:HGNC Symbol;Acc:HGNC:11776] | 1668 | 1.039 | -0.0036 | No |
| 4 | PIGN | phosphatidylinositol glycan anchor biosynthesis class N [Source:HGNC Symbol;Acc:HGNC:8967] | 1808 | 0.982 | 0.0044 | No |
| 5 | DLL1 | delta like canonical Notch ligand 1 [Source:HGNC Symbol;Acc:HGNC:2908] | 2759 | 0.648 | -0.0136 | No |
| 6 | FANCL | FA complementation group L [Source:HGNC Symbol;Acc:HGNC:20748] | 3669 | 0.448 | -0.0328 | No |
| 7 | FGFR1 | fibroblast growth factor receptor 1 [Source:HGNC Symbol;Acc:HGNC:3688] | 4566 | 0.299 | -0.0534 | No |
| 8 | RBM10 | RNA binding motif protein 10 [Source:HGNC Symbol;Acc:HGNC:9896] | 5858 | 0.105 | -0.0870 | No |
| 9 | SLX4 | SLX4 structure-specific endonuclease subunit [Source:HGNC Symbol;Acc:HGNC:23845] | 5984 | 0.089 | -0.0893 | No |
| 10 | EP300 | E1A binding protein p300 [Source:HGNC Symbol;Acc:HGNC:3373] | 6373 | 0.043 | -0.0993 | No |
| 11 | DYRK1A | dual specificity tyrosine phosphorylation regulated kinase 1A [Source:HGNC Symbol;Acc:HGNC:3091] | 6418 | 0.037 | -0.1001 | No |
| 12 | FANCB | FA complementation group B [Source:HGNC Symbol;Acc:HGNC:3583] | 6595 | 0.016 | -0.1046 | No |
| 13 | FBN2 | fibrillin 2 [Source:HGNC Symbol;Acc:HGNC:3604] | 8537 | 0.000 | -0.1571 | No |
| 14 | SIX3 | SIX homeobox 3 [Source:HGNC Symbol;Acc:HGNC:10889] | 8769 | 0.000 | -0.1633 | No |
| 15 | CDKN1A | cyclin dependent kinase inhibitor 1A [Source:HGNC Symbol;Acc:HGNC:1784] | 12854 | 0.000 | -0.2736 | No |
| 16 | GAS1 | growth arrest specific 1 [Source:HGNC Symbol;Acc:HGNC:4165] | 12868 | 0.000 | -0.2740 | No |
| 17 | CDKN2B | cyclin dependent kinase inhibitor 2B [Source:HGNC Symbol;Acc:HGNC:1788] | 12877 | 0.000 | -0.2742 | No |
| 18 | GATA1 | GATA binding protein 1 [Source:HGNC Symbol;Acc:HGNC:4170] | 13737 | 0.000 | -0.2974 | No |
| 19 | SALL1 | spalt like transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:10524] | 14195 | 0.000 | -0.3098 | No |
| 20 | RFX6 | regulatory factor X6 [Source:HGNC Symbol;Acc:HGNC:21478] | 14564 | 0.000 | -0.3197 | No |
| 21 | STRA6 | signaling receptor and transporter of retinol STRA6 [Source:HGNC Symbol;Acc:HGNC:30650] | 16583 | 0.000 | -0.3742 | No |
| 22 | FANCF | FA complementation group F [Source:HGNC Symbol;Acc:HGNC:3587] | 18490 | 0.000 | -0.4257 | No |
| 23 | PLG | plasminogen [Source:HGNC Symbol;Acc:HGNC:9071] | 20029 | 0.000 | -0.4673 | No |
| 24 | SHH | sonic hedgehog signaling molecule [Source:HGNC Symbol;Acc:HGNC:10848] | 20840 | 0.000 | -0.4892 | No |
| 25 | GLI2 | GLI family zinc finger 2 [Source:HGNC Symbol;Acc:HGNC:4318] | 21764 | 0.000 | -0.5141 | No |
| 26 | FOXH1 | forkhead box H1 [Source:HGNC Symbol;Acc:HGNC:3814] | 23632 | 0.000 | -0.5645 | No |
| 27 | FOXF1 | forkhead box F1 [Source:HGNC Symbol;Acc:HGNC:3809] | 23653 | 0.000 | -0.5651 | No |
| 28 | TDGF1 | teratocarcinoma-derived growth factor 1 [Source:HGNC Symbol;Acc:HGNC:11701] | 24698 | 0.000 | -0.5933 | No |
| 29 | FGF8 | fibroblast growth factor 8 [Source:HGNC Symbol;Acc:HGNC:3686] | 25427 | 0.000 | -0.6130 | No |
| 30 | CARD8 | caspase recruitment domain family member 8 [Source:HGNC Symbol;Acc:HGNC:17057] | 26078 | 0.000 | -0.6305 | No |
| 31 | BUB3 | BUB3 mitotic checkpoint protein [Source:HGNC Symbol;Acc:HGNC:1151] | 28342 | -0.112 | -0.6903 | No |
| 32 | NODAL | nodal growth differentiation factor [Source:HGNC Symbol;Acc:HGNC:7865] | 28655 | -0.146 | -0.6970 | No |
| 33 | STAG2 | stromal antigen 2 [Source:HGNC Symbol;Acc:HGNC:11355] | 29511 | -0.253 | -0.7171 | No |
| 34 | XRCC2 | X-ray repair cross complementing 2 [Source:HGNC Symbol;Acc:HGNC:12829] | 30026 | -0.316 | -0.7272 | No |
| 35 | ADAM17 | ADAM metallopeptidase domain 17 [Source:HGNC Symbol;Acc:HGNC:195] | 30244 | -0.344 | -0.7289 | No |
| 36 | ERCC4 | ERCC excision repair 4, endonuclease catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3436] | 30540 | -0.384 | -0.7323 | No |
| 37 | PLA2G4A | phospholipase A2 group IVA [Source:HGNC Symbol;Acc:HGNC:9035] | 31188 | -0.482 | -0.7440 | No |
| 38 | FANCC | FA complementation group C [Source:HGNC Symbol;Acc:HGNC:3584] | 31450 | -0.521 | -0.7449 | No |
| 39 | FANCG | FA complementation group G [Source:HGNC Symbol;Acc:HGNC:3588] | 32511 | -0.724 | -0.7648 | Yes |
| 40 | BRIP1 | BRCA1 interacting protein C-terminal helicase 1 [Source:HGNC Symbol;Acc:HGNC:20473] | 32642 | -0.756 | -0.7593 | Yes |
| 41 | SUFU | SUFU negative regulator of hedgehog signaling [Source:HGNC Symbol;Acc:HGNC:16466] | 32660 | -0.760 | -0.7507 | Yes |
| 42 | CDON | cell adhesion associated, oncogene regulated [Source:HGNC Symbol;Acc:HGNC:17104] | 32832 | -0.800 | -0.7457 | Yes |
| 43 | DISP1 | dispatched RND transporter family member 1 [Source:HGNC Symbol;Acc:HGNC:19711] | 32853 | -0.805 | -0.7367 | Yes |
| 44 | MEN1 | menin 1 [Source:HGNC Symbol;Acc:HGNC:7010] | 33085 | -0.862 | -0.7326 | Yes |
| 45 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33264 | -0.912 | -0.7265 | Yes |
| 46 | RFWD3 | ring finger and WD repeat domain 3 [Source:HGNC Symbol;Acc:HGNC:25539] | 33487 | -0.970 | -0.7209 | Yes |
| 47 | FANCM | FA complementation group M [Source:HGNC Symbol;Acc:HGNC:23168] | 33771 | -1.047 | -0.7160 | Yes |
| 48 | FANCA | FA complementation group A [Source:HGNC Symbol;Acc:HGNC:3582] | 33866 | -1.073 | -0.7057 | Yes |
| 49 | RTTN | rotatin [Source:HGNC Symbol;Acc:HGNC:18654] | 34015 | -1.119 | -0.6964 | Yes |
| 50 | CDC45 | cell division cycle 45 [Source:HGNC Symbol;Acc:HGNC:1739] | 34224 | -1.190 | -0.6877 | Yes |
| 51 | MAD2L2 | mitotic arrest deficient 2 like 2 [Source:HGNC Symbol;Acc:HGNC:6764] | 34439 | -1.267 | -0.6784 | Yes |
| 52 | CHST14 | carbohydrate sulfotransferase 14 [Source:HGNC Symbol;Acc:HGNC:24464] | 34504 | -1.293 | -0.6646 | Yes |
| 53 | FANCE | FA complementation group E [Source:HGNC Symbol;Acc:HGNC:3586] | 34609 | -1.335 | -0.6515 | Yes |
| 54 | KDM3B | lysine demethylase 3B [Source:HGNC Symbol;Acc:HGNC:1337] | 35112 | -1.574 | -0.6462 | Yes |
| 55 | BRCA2 | BRCA2 DNA repair associated [Source:HGNC Symbol;Acc:HGNC:1101] | 35234 | -1.641 | -0.6299 | Yes |
| 56 | RAD21 | RAD21 cohesin complex component [Source:HGNC Symbol;Acc:HGNC:9811] | 35280 | -1.666 | -0.6112 | Yes |
| 57 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35320 | -1.686 | -0.5921 | Yes |
| 58 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35401 | -1.734 | -0.5735 | Yes |
| 59 | CEP57 | centrosomal protein 57 [Source:HGNC Symbol;Acc:HGNC:30794] | 35418 | -1.743 | -0.5531 | Yes |
| 60 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 35435 | -1.752 | -0.5326 | Yes |
| 61 | RERE | arginine-glutamic acid dipeptide repeats [Source:HGNC Symbol;Acc:HGNC:9965] | 35742 | -1.956 | -0.5175 | Yes |
| 62 | UBE2T | ubiquitin conjugating enzyme E2 T [Source:HGNC Symbol;Acc:HGNC:25009] | 36026 | -2.171 | -0.4991 | Yes |
| 63 | PTCH1 | patched 1 [Source:HGNC Symbol;Acc:HGNC:9585] | 36066 | -2.205 | -0.4738 | Yes |
| 64 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 36081 | -2.221 | -0.4477 | Yes |
| 65 | PORCN | porcupine O-acyltransferase [Source:HGNC Symbol;Acc:HGNC:17652] | 36099 | -2.239 | -0.4213 | Yes |
| 66 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 36101 | -2.242 | -0.3946 | Yes |
| 67 | CDKN1B | cyclin dependent kinase inhibitor 1B [Source:HGNC Symbol;Acc:HGNC:1785] | 36244 | -2.370 | -0.3701 | Yes |
| 68 | ARID1B | AT-rich interaction domain 1B [Source:HGNC Symbol;Acc:HGNC:18040] | 36294 | -2.422 | -0.3424 | Yes |
| 69 | MYCN | MYCN proto-oncogene, bHLH transcription factor [Source:HGNC Symbol;Acc:HGNC:7559] | 36371 | -2.507 | -0.3145 | Yes |
| 70 | FLI1 | Fli-1 proto-oncogene, ETS transcription factor [Source:HGNC Symbol;Acc:HGNC:3749] | 36425 | -2.582 | -0.2851 | Yes |
| 71 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36453 | -2.630 | -0.2544 | Yes |
| 72 | FANCD2 | FA complementation group D2 [Source:HGNC Symbol;Acc:HGNC:3585] | 36504 | -2.685 | -0.2236 | Yes |
| 73 | PALB2 | partner and localizer of BRCA2 [Source:HGNC Symbol;Acc:HGNC:26144] | 36542 | -2.740 | -0.1918 | Yes |
| 74 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 36644 | -2.890 | -0.1600 | Yes |
| 75 | BRCA1 | BRCA1 DNA repair associated [Source:HGNC Symbol;Acc:HGNC:1100] | 36790 | -3.161 | -0.1261 | Yes |
| 76 | ZIC2 | Zic family member 2 [Source:HGNC Symbol;Acc:HGNC:12873] | 36793 | -3.167 | -0.0883 | Yes |
| 77 | TTC7A | tetratricopeptide repeat domain 7A [Source:HGNC Symbol;Acc:HGNC:19750] | 36984 | -3.790 | -0.0482 | Yes |
| 78 | RARB | retinoic acid receptor beta [Source:HGNC Symbol;Acc:HGNC:9865] | 37057 | -4.265 | 0.0009 | Yes |