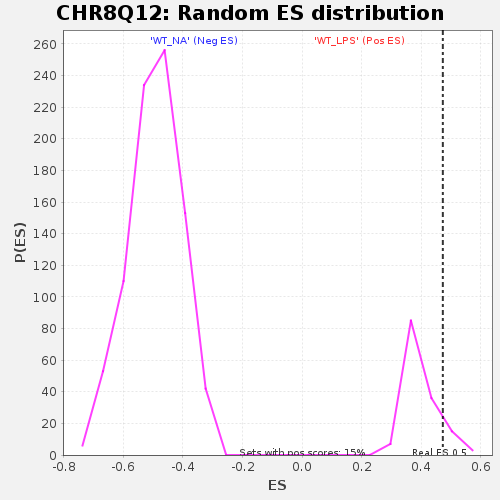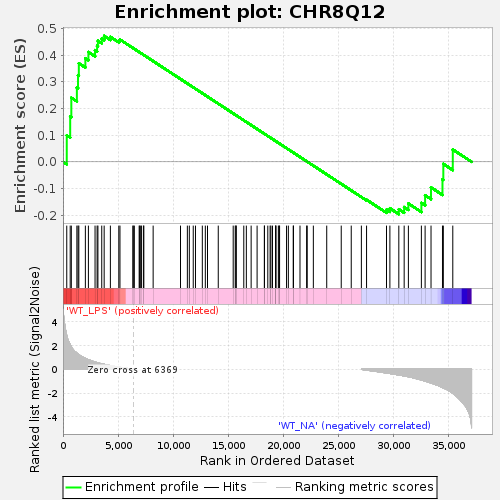
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_LPS_versus_WT_NA.greenbergcls.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_LPS |
| GeneSet | CHR8Q12 |
| Enrichment Score (ES) | 0.47200406 |
| Normalized Enrichment Score (NES) | 1.1877806 |
| Nominal p-value | 0.11643836 |
| FDR q-value | 0.6195556 |
| FWER p-Value | 0.98 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | LYN | LYN proto-oncogene, Src family tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:6735] | 341 | 2.816 | 0.0992 | Yes |
| 2 | PDCL3P1 | PDCL3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31826] | 653 | 2.064 | 0.1703 | Yes |
| 3 | TMEM68 | transmembrane protein 68 [Source:HGNC Symbol;Acc:HGNC:26510] | 756 | 1.888 | 0.2402 | Yes |
| 4 | NARS1P2 | asparaginyl-tRNA synthetase 1 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:50481] | 1259 | 1.343 | 0.2784 | Yes |
| 5 | CHD7 | chromodomain helicase DNA binding protein 7 [Source:HGNC Symbol;Acc:HGNC:20626] | 1364 | 1.270 | 0.3245 | Yes |
| 6 | TGS1 | trimethylguanosine synthase 1 [Source:HGNC Symbol;Acc:HGNC:17843] | 1437 | 1.214 | 0.3693 | Yes |
| 7 | YTHDF3 | YTH N6-methyladenosine RNA binding protein 3 [Source:HGNC Symbol;Acc:HGNC:26465] | 2030 | 0.907 | 0.3882 | Yes |
| 8 | SDCBP | syndecan binding protein [Source:HGNC Symbol;Acc:HGNC:10662] | 2306 | 0.797 | 0.4114 | Yes |
| 9 | PENK | proenkephalin [Source:HGNC Symbol;Acc:HGNC:8831] | 2909 | 0.604 | 0.4184 | Yes |
| 10 | RAB2A | RAB2A, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9763] | 3096 | 0.559 | 0.4349 | Yes |
| 11 | RPS20 | ribosomal protein S20 [Source:HGNC Symbol;Acc:HGNC:10405] | 3170 | 0.541 | 0.4537 | Yes |
| 12 | RPL30P10 | ribosomal protein L30 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:36822] | 3517 | 0.457 | 0.4620 | Yes |
| 13 | IFITM8P | interferon induced transmembrane protein 8 pseudogene [Source:HGNC Symbol;Acc:HGNC:32202] | 3737 | 0.413 | 0.4720 | Yes |
| 14 | NPM1P6 | nucleophosmin 1 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:7926] | 4301 | 0.302 | 0.4684 | No |
| 15 | NPM1P21 | nucleophosmin 1 pseudogene 21 [Source:HGNC Symbol;Acc:HGNC:13855] | 5065 | 0.183 | 0.4549 | No |
| 16 | CHCHD7 | coiled-coil-helix-coiled-coil-helix domain containing 7 [Source:HGNC Symbol;Acc:HGNC:28314] | 5171 | 0.166 | 0.4584 | No |
| 17 | UBXN2B | UBX domain protein 2B [Source:HGNC Symbol;Acc:HGNC:27035] | 6340 | 0.003 | 0.4270 | No |
| 18 | AC009597.1 | novel transcript | 6381 | 0.000 | 0.4259 | No |
| 19 | RNU6-596P | RNA, U6 small nuclear 596, pseudogene [Source:HGNC Symbol;Acc:HGNC:47559] | 6476 | 0.000 | 0.4234 | No |
| 20 | AC012349.1 | novel transcript | 6914 | 0.000 | 0.4116 | No |
| 21 | AC011124.2 | novel transcript | 6959 | 0.000 | 0.4104 | No |
| 22 | LINC01301 | long intergenic non-protein coding RNA 1301 [Source:HGNC Symbol;Acc:HGNC:50464] | 7053 | 0.000 | 0.4079 | No |
| 23 | LINC01289 | long intergenic non-protein coding RNA 1289 [Source:HGNC Symbol;Acc:HGNC:50354] | 7135 | 0.000 | 0.4057 | No |
| 24 | LINC01606 | long intergenic non-protein coding RNA 1606 [Source:HGNC Symbol;Acc:HGNC:51656] | 7321 | 0.000 | 0.4007 | No |
| 25 | LINC01602 | long intergenic non-protein coding RNA 1602 [Source:HGNC Symbol;Acc:HGNC:51634] | 7329 | 0.000 | 0.4005 | No |
| 26 | BHLHE22 | basic helix-loop-helix family member e22 [Source:HGNC Symbol;Acc:HGNC:11963] | 8183 | 0.000 | 0.3774 | No |
| 27 | KRT8P3 | keratin 8 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:31056] | 10667 | 0.000 | 0.3104 | No |
| 28 | AC092819.1 | novel transcript | 11289 | 0.000 | 0.2936 | No |
| 29 | RNU4-50P | RNA, U4 small nuclear 50, pseudogene [Source:HGNC Symbol;Acc:HGNC:46986] | 11467 | 0.000 | 0.2888 | No |
| 30 | AC068075.1 | novel transcript | 11823 | 0.000 | 0.2792 | No |
| 31 | LINC00968 | long intergenic non-protein coding RNA 968 [Source:HGNC Symbol;Acc:HGNC:48727] | 12026 | 0.000 | 0.2737 | No |
| 32 | TOX | thymocyte selection associated high mobility group box [Source:HGNC Symbol;Acc:HGNC:18988] | 12647 | 0.000 | 0.2570 | No |
| 33 | AC090200.1 | novel transcript | 12920 | 0.000 | 0.2496 | No |
| 34 | AC104051.2 | novel transcript | 13118 | 0.000 | 0.2443 | No |
| 35 | LINC02155 | long intergenic non-protein coding RNA 2155 [Source:HGNC Symbol;Acc:HGNC:53016] | 14094 | 0.000 | 0.2180 | No |
| 36 | COX6CP8 | cytochrome c oxidase subunit 6C pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:49360] | 15450 | 0.000 | 0.1814 | No |
| 37 | NKAIN3 | sodium/potassium transporting ATPase interacting 3 [Source:HGNC Symbol;Acc:HGNC:26829] | 15635 | 0.000 | 0.1764 | No |
| 38 | CA8 | carbonic anhydrase 8 [Source:HGNC Symbol;Acc:HGNC:1382] | 15701 | 0.000 | 0.1746 | No |
| 39 | CLVS1 | clavesin 1 [Source:HGNC Symbol;Acc:HGNC:23139] | 15725 | 0.000 | 0.1740 | No |
| 40 | CERNA3 | competing endogenous lncRNA 3 for miR-645 [Source:HGNC Symbol;Acc:HGNC:53469] | 16411 | 0.000 | 0.1555 | No |
| 41 | RN7SL323P | RNA, 7SL, cytoplasmic 323, pseudogene [Source:HGNC Symbol;Acc:HGNC:46339] | 16644 | 0.000 | 0.1492 | No |
| 42 | AC090136.3 | novel transcript | 17079 | 0.000 | 0.1375 | No |
| 43 | RN7SL798P | RNA, 7SL, cytoplasmic 798, pseudogene [Source:HGNC Symbol;Acc:HGNC:46814] | 17620 | 0.000 | 0.1229 | No |
| 44 | XKR4 | XK related 4 [Source:HGNC Symbol;Acc:HGNC:29394] | 18281 | 0.000 | 0.1051 | No |
| 45 | RN7SL135P | RNA, 7SL, cytoplasmic 135, pseudogene [Source:HGNC Symbol;Acc:HGNC:46151] | 18287 | 0.000 | 0.1050 | No |
| 46 | RN7SKP97 | RN7SK pseudogene 97 [Source:HGNC Symbol;Acc:HGNC:45821] | 18591 | 0.000 | 0.0968 | No |
| 47 | MIR124-2 | microRNA 124-2 [Source:HGNC Symbol;Acc:HGNC:31503] | 18801 | 0.000 | 0.0911 | No |
| 48 | MOS | MOS proto-oncogene, serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:7199] | 18874 | 0.000 | 0.0892 | No |
| 49 | YTHDF3-AS1 | YTHDF3 antisense RNA 1 (head to head) [Source:HGNC Symbol;Acc:HGNC:48728] | 19017 | 0.000 | 0.0853 | No |
| 50 | RNA5SP267 | RNA, 5S ribosomal pseudogene 267 [Source:HGNC Symbol;Acc:HGNC:43167] | 19313 | 0.000 | 0.0774 | No |
| 51 | RNA5SP266 | RNA, 5S ribosomal pseudogene 266 [Source:HGNC Symbol;Acc:HGNC:43166] | 19314 | 0.000 | 0.0774 | No |
| 52 | RNA5SP265 | RNA, 5S ribosomal pseudogene 265 [Source:HGNC Symbol;Acc:HGNC:43165] | 19315 | 0.000 | 0.0774 | No |
| 53 | C1GALT1P3 | C1GALT1 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:51616] | 19550 | 0.000 | 0.0711 | No |
| 54 | AC090152.1 | novel transcript | 19620 | 0.000 | 0.0692 | No |
| 55 | MIR124-2HG | MIR124-2 host gene [Source:HGNC Symbol;Acc:HGNC:48723] | 19661 | 0.000 | 0.0681 | No |
| 56 | RN7SKP135 | RN7SK pseudogene 135 [Source:HGNC Symbol;Acc:HGNC:45859] | 20293 | 0.000 | 0.0511 | No |
| 57 | SLC2A13P1 | SLC2A13 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:48929] | 20457 | 0.000 | 0.0467 | No |
| 58 | CYP7B1 | cytochrome P450 family 7 subfamily B member 1 [Source:HGNC Symbol;Acc:HGNC:2652] | 20900 | 0.000 | 0.0347 | No |
| 59 | CYP7A1 | cytochrome P450 family 7 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2651] | 20923 | 0.000 | 0.0341 | No |
| 60 | AC021393.1 | uncharacterized LOC105375861 [Source:NCBI gene (formerly Entrezgene);Acc:105375861] | 21514 | 0.000 | 0.0182 | No |
| 61 | SNORD54 | small nucleolar RNA, C/D box 54 [Source:HGNC Symbol;Acc:HGNC:10204] | 22116 | 0.000 | 0.0019 | No |
| 62 | TTPA | alpha tocopherol transfer protein [Source:HGNC Symbol;Acc:HGNC:12404] | 22168 | 0.000 | 0.0006 | No |
| 63 | RNU6-13P | RNA, U6 small nuclear 13, pseudogene [Source:HGNC Symbol;Acc:HGNC:34257] | 22726 | 0.000 | -0.0145 | No |
| 64 | LINC00588 | long intergenic non-protein coding RNA 588 [Source:HGNC Symbol;Acc:HGNC:24494] | 23941 | 0.000 | -0.0473 | No |
| 65 | MIR4470 | microRNA 4470 [Source:HGNC Symbol;Acc:HGNC:41833] | 25258 | 0.000 | -0.0828 | No |
| 66 | SDR16C6P | short chain dehydrogenase/reductase family 16C member 6, pseudogene [Source:HGNC Symbol;Acc:HGNC:35413] | 26166 | 0.000 | -0.1073 | No |
| 67 | SDR16C5 | short chain dehydrogenase/reductase family 16C member 5 [Source:HGNC Symbol;Acc:HGNC:30311] | 27086 | 0.000 | -0.1322 | No |
| 68 | GGH | gamma-glutamyl hydrolase [Source:HGNC Symbol;Acc:HGNC:4248] | 27557 | -0.067 | -0.1423 | No |
| 69 | RPL37P6 | ribosomal protein L37 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:31080] | 29368 | -0.308 | -0.1794 | No |
| 70 | RPL26P26 | ribosomal protein L26 pseudogene 26 [Source:HGNC Symbol;Acc:HGNC:35693] | 29675 | -0.350 | -0.1742 | No |
| 71 | FAM110B | family with sequence similarity 110 member B [Source:HGNC Symbol;Acc:HGNC:28587] | 30485 | -0.469 | -0.1780 | No |
| 72 | ASPH | aspartate beta-hydroxylase [Source:HGNC Symbol;Acc:HGNC:757] | 30971 | -0.551 | -0.1698 | No |
| 73 | BPNT2 | 3'(2'), 5'-bisphosphate nucleotidase 2 [Source:HGNC Symbol;Acc:HGNC:26019] | 31354 | -0.616 | -0.1564 | No |
| 74 | SBF1P1 | SET binding factor 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:31098] | 32533 | -0.887 | -0.1541 | No |
| 75 | PLAG1 | PLAG1 zinc finger [Source:HGNC Symbol;Acc:HGNC:9045] | 32872 | -0.980 | -0.1255 | No |
| 76 | XRCC6P4 | X-ray repair cross complementing 6 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:45186] | 33409 | -1.141 | -0.0961 | No |
| 77 | SRPK2P | SRSF protein kinase 2 pseudogene [Source:HGNC Symbol;Acc:HGNC:30303] | 34449 | -1.521 | -0.0656 | No |
| 78 | NSMAF | neutral sphingomyelinase activation associated factor [Source:HGNC Symbol;Acc:HGNC:8017] | 34526 | -1.551 | -0.0079 | No |
| 79 | SEPTIN10P1 | septin 10 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:40018] | 35385 | -2.004 | 0.0461 | No |