Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_LPS_versus_WT_NA.greenbergcls.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | CROONQUIST_IL6_DEPRIVATION_DN |
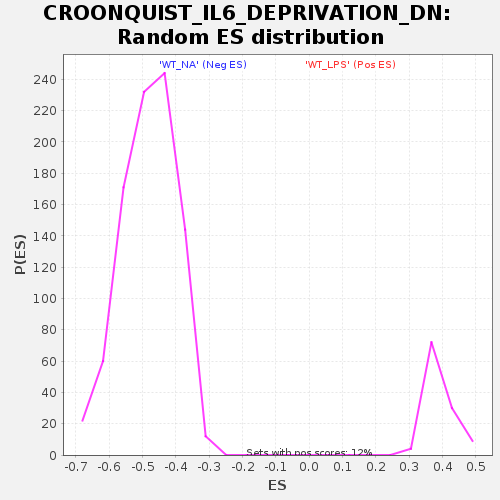
| Enrichment Score (ES) | -0.8169364 |
| Normalized Enrichment Score (NES) | -1.7005903 |
| Nominal p-value | 0.0 |
| FDR q-value | 5.6608984E-5 |
| FWER p-Value | 0.001 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SPP1 | secreted phosphoprotein 1 [Source:HGNC Symbol;Acc:HGNC:11255] | 180 | 3.595 | 0.0125 | No |
| 2 | ABHD5 | abhydrolase domain containing 5, lysophosphatidic acid acyltransferase [Source:HGNC Symbol;Acc:HGNC:21396] | 877 | 1.691 | 0.0018 | No |
| 3 | RRP12 | ribosomal RNA processing 12 homolog [Source:HGNC Symbol;Acc:HGNC:29100] | 1081 | 1.488 | 0.0035 | No |
| 4 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 1268 | 1.337 | 0.0049 | No |
| 5 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 1626 | 1.102 | 0.0006 | No |
| 6 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1902 | 0.962 | -0.0022 | No |
| 7 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 3064 | 0.566 | -0.0308 | No |
| 8 | PTPRC | protein tyrosine phosphatase receptor type C [Source:HGNC Symbol;Acc:HGNC:9666] | 3218 | 0.526 | -0.0324 | No |
| 9 | MYRF | myelin regulatory factor [Source:HGNC Symbol;Acc:HGNC:1181] | 3781 | 0.406 | -0.0456 | No |
| 10 | CDC25A | cell division cycle 25A [Source:HGNC Symbol;Acc:HGNC:1725] | 4007 | 0.358 | -0.0500 | No |
| 11 | DHFR | dihydrofolate reductase [Source:HGNC Symbol;Acc:HGNC:2861] | 6236 | 0.018 | -0.1101 | No |
| 12 | AASS | aminoadipate-semialdehyde synthase [Source:HGNC Symbol;Acc:HGNC:17366] | 10442 | 0.000 | -0.2238 | No |
| 13 | APOBEC3B | apolipoprotein B mRNA editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] | 12467 | 0.000 | -0.2785 | No |
| 14 | FRZB | frizzled related protein [Source:HGNC Symbol;Acc:HGNC:3959] | 16691 | 0.000 | -0.3927 | No |
| 15 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 25804 | 0.000 | -0.6390 | No |
| 16 | GGH | gamma-glutamyl hydrolase [Source:HGNC Symbol;Acc:HGNC:4248] | 27557 | -0.067 | -0.6860 | No |
| 17 | ITM2A | integral membrane protein 2A [Source:HGNC Symbol;Acc:HGNC:6173] | 28372 | -0.169 | -0.7072 | No |
| 18 | ANP32E | acidic nuclear phosphoprotein 32 family member E [Source:HGNC Symbol;Acc:HGNC:16673] | 31243 | -0.597 | -0.7819 | No |
| 19 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 31272 | -0.602 | -0.7797 | No |
| 20 | GMPS | guanine monophosphate synthase [Source:HGNC Symbol;Acc:HGNC:4378] | 31457 | -0.639 | -0.7816 | No |
| 21 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 31585 | -0.666 | -0.7818 | No |
| 22 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32856 | -0.976 | -0.8114 | No |
| 23 | SPOUT1 | SPOUT domain containing methyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:26933] | 33043 | -1.031 | -0.8115 | No |
| 24 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33245 | -1.087 | -0.8117 | Yes |
| 25 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33258 | -1.093 | -0.8067 | Yes |
| 26 | CDC45 | cell division cycle 45 [Source:HGNC Symbol;Acc:HGNC:1739] | 33394 | -1.135 | -0.8049 | Yes |
| 27 | SEPHS1 | selenophosphate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:19685] | 33457 | -1.155 | -0.8010 | Yes |
| 28 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33793 | -1.266 | -0.8040 | Yes |
| 29 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 33887 | -1.300 | -0.8002 | Yes |
| 30 | KNTC1 | kinetochore associated 1 [Source:HGNC Symbol;Acc:HGNC:17255] | 33893 | -1.302 | -0.7940 | Yes |
| 31 | H2BC11 | H2B clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4761] | 33907 | -1.308 | -0.7881 | Yes |
| 32 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 34101 | -1.375 | -0.7867 | Yes |
| 33 | SKP2 | S-phase kinase associated protein 2 [Source:HGNC Symbol;Acc:HGNC:10901] | 34311 | -1.461 | -0.7853 | Yes |
| 34 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34588 | -1.576 | -0.7851 | Yes |
| 35 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34938 | -1.741 | -0.7862 | Yes |
| 36 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35043 | -1.792 | -0.7803 | Yes |
| 37 | KPNA2 | karyopherin subunit alpha 2 [Source:HGNC Symbol;Acc:HGNC:6395] | 35063 | -1.804 | -0.7721 | Yes |
| 38 | CHAF1A | chromatin assembly factor 1 subunit A [Source:HGNC Symbol;Acc:HGNC:1910] | 35073 | -1.810 | -0.7636 | Yes |
| 39 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 35148 | -1.850 | -0.7567 | Yes |
| 40 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 35386 | -2.005 | -0.7534 | Yes |
| 41 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35573 | -2.135 | -0.7482 | Yes |
| 42 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35579 | -2.138 | -0.7380 | Yes |
| 43 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35675 | -2.217 | -0.7299 | Yes |
| 44 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 35717 | -2.250 | -0.7201 | Yes |
| 45 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 35735 | -2.266 | -0.7096 | Yes |
| 46 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35799 | -2.315 | -0.7002 | Yes |
| 47 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35816 | -2.333 | -0.6893 | Yes |
| 48 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 35837 | -2.347 | -0.6785 | Yes |
| 49 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35874 | -2.384 | -0.6680 | Yes |
| 50 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 35897 | -2.409 | -0.6570 | Yes |
| 51 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 35899 | -2.411 | -0.6454 | Yes |
| 52 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35901 | -2.411 | -0.6338 | Yes |
| 53 | MAD2L1 | mitotic arrest deficient 2 like 1 [Source:HGNC Symbol;Acc:HGNC:6763] | 35937 | -2.441 | -0.6229 | Yes |
| 54 | STIL | STIL centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:10879] | 35955 | -2.459 | -0.6115 | Yes |
| 55 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36076 | -2.578 | -0.6023 | Yes |
| 56 | POLE2 | DNA polymerase epsilon 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9178] | 36099 | -2.607 | -0.5903 | Yes |
| 57 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 36108 | -2.616 | -0.5779 | Yes |
| 58 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 36127 | -2.643 | -0.5657 | Yes |
| 59 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36139 | -2.657 | -0.5531 | Yes |
| 60 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 36141 | -2.658 | -0.5403 | Yes |
| 61 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36164 | -2.684 | -0.5280 | Yes |
| 62 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 36261 | -2.777 | -0.5172 | Yes |
| 63 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 36279 | -2.795 | -0.5041 | Yes |
| 64 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 36332 | -2.865 | -0.4917 | Yes |
| 65 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36339 | -2.870 | -0.4780 | Yes |
| 66 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36371 | -2.891 | -0.4649 | Yes |
| 67 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36383 | -2.908 | -0.4512 | Yes |
| 68 | GALK1 | galactokinase 1 [Source:HGNC Symbol;Acc:HGNC:4118] | 36447 | -2.976 | -0.4385 | Yes |
| 69 | TRAIP | TRAF interacting protein [Source:HGNC Symbol;Acc:HGNC:30764] | 36449 | -2.976 | -0.4242 | Yes |
| 70 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 36483 | -3.006 | -0.4106 | Yes |
| 71 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36491 | -3.015 | -0.3962 | Yes |
| 72 | KIF14 | kinesin family member 14 [Source:HGNC Symbol;Acc:HGNC:19181] | 36494 | -3.018 | -0.3817 | Yes |
| 73 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36555 | -3.099 | -0.3684 | Yes |
| 74 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36592 | -3.140 | -0.3542 | Yes |
| 75 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36611 | -3.176 | -0.3394 | Yes |
| 76 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36630 | -3.207 | -0.3244 | Yes |
| 77 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 36665 | -3.258 | -0.3096 | Yes |
| 78 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36667 | -3.270 | -0.2938 | Yes |
| 79 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36680 | -3.288 | -0.2783 | Yes |
| 80 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 36692 | -3.321 | -0.2625 | Yes |
| 81 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36707 | -3.354 | -0.2467 | Yes |
| 82 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 36709 | -3.356 | -0.2306 | Yes |
| 83 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 36724 | -3.390 | -0.2146 | Yes |
| 84 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 36770 | -3.470 | -0.1990 | Yes |
| 85 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36833 | -3.603 | -0.1833 | Yes |
| 86 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 36862 | -3.659 | -0.1664 | Yes |
| 87 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 36868 | -3.675 | -0.1488 | Yes |
| 88 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36876 | -3.698 | -0.1312 | Yes |
| 89 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36891 | -3.747 | -0.1135 | Yes |
| 90 | BRCA1 | BRCA1 DNA repair associated [Source:HGNC Symbol;Acc:HGNC:1100] | 36897 | -3.766 | -0.0954 | Yes |
| 91 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36917 | -3.817 | -0.0775 | Yes |
| 92 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36955 | -3.967 | -0.0594 | Yes |
| 93 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 36989 | -4.089 | -0.0405 | Yes |
| 94 | CCNE2 | cyclin E2 [Source:HGNC Symbol;Acc:HGNC:1590] | 37055 | -4.466 | -0.0207 | Yes |
| 95 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37060 | -4.490 | 0.0008 | Yes |