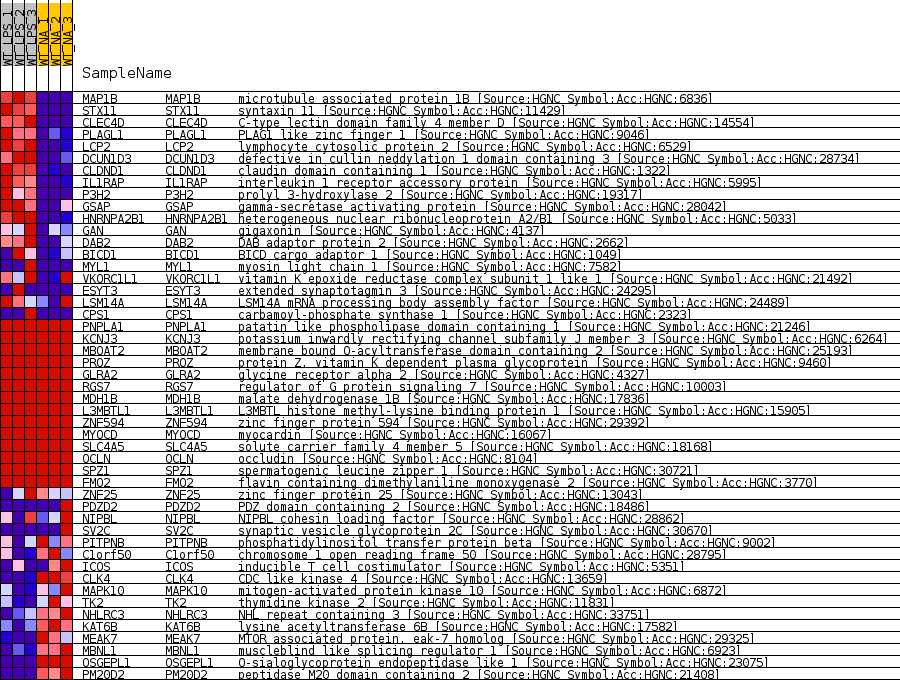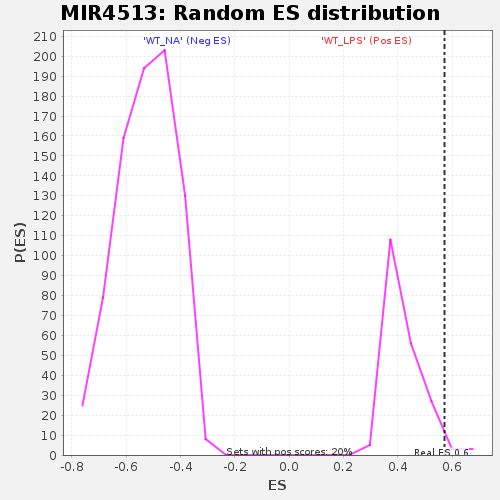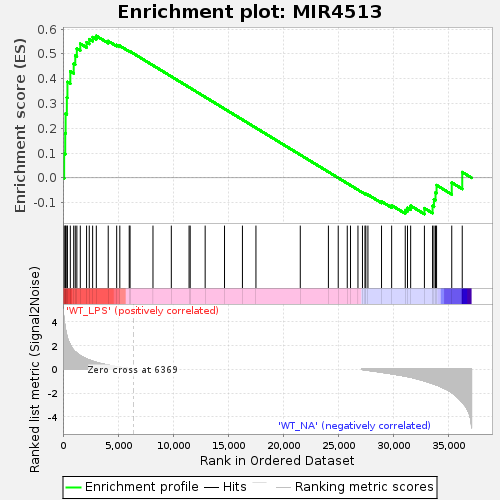
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_LPS_versus_WT_NA.greenbergcls.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_LPS |
| GeneSet | MIR4513 |
| Enrichment Score (ES) | 0.5721888 |
| Normalized Enrichment Score (NES) | 1.3566716 |
| Nominal p-value | 0.024752475 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MAP1B | microtubule associated protein 1B [Source:HGNC Symbol;Acc:HGNC:6836] | 104 | 4.074 | 0.0971 | Yes |
| 2 | STX11 | syntaxin 11 [Source:HGNC Symbol;Acc:HGNC:11429] | 202 | 3.471 | 0.1796 | Yes |
| 3 | CLEC4D | C-type lectin domain family 4 member D [Source:HGNC Symbol;Acc:HGNC:14554] | 236 | 3.282 | 0.2592 | Yes |
| 4 | PLAGL1 | PLAG1 like zinc finger 1 [Source:HGNC Symbol;Acc:HGNC:9046] | 349 | 2.772 | 0.3242 | Yes |
| 5 | LCP2 | lymphocyte cytosolic protein 2 [Source:HGNC Symbol;Acc:HGNC:6529] | 402 | 2.598 | 0.3865 | Yes |
| 6 | DCUN1D3 | defective in cullin neddylation 1 domain containing 3 [Source:HGNC Symbol;Acc:HGNC:28734] | 664 | 2.041 | 0.4295 | Yes |
| 7 | CLDND1 | claudin domain containing 1 [Source:HGNC Symbol;Acc:HGNC:1322] | 972 | 1.585 | 0.4601 | Yes |
| 8 | IL1RAP | interleukin 1 receptor accessory protein [Source:HGNC Symbol;Acc:HGNC:5995] | 1113 | 1.464 | 0.4922 | Yes |
| 9 | P3H2 | prolyl 3-hydroxylase 2 [Source:HGNC Symbol;Acc:HGNC:19317] | 1258 | 1.343 | 0.5213 | Yes |
| 10 | GSAP | gamma-secretase activating protein [Source:HGNC Symbol;Acc:HGNC:28042] | 1570 | 1.138 | 0.5408 | Yes |
| 11 | HNRNPA2B1 | heterogeneous nuclear ribonucleoprotein A2/B1 [Source:HGNC Symbol;Acc:HGNC:5033] | 2140 | 0.862 | 0.5466 | Yes |
| 12 | GAN | gigaxonin [Source:HGNC Symbol;Acc:HGNC:4137] | 2385 | 0.769 | 0.5588 | Yes |
| 13 | DAB2 | DAB adaptor protein 2 [Source:HGNC Symbol;Acc:HGNC:2662] | 2690 | 0.671 | 0.5671 | Yes |
| 14 | BICD1 | BICD cargo adaptor 1 [Source:HGNC Symbol;Acc:HGNC:1049] | 3025 | 0.575 | 0.5722 | Yes |
| 15 | MYL1 | myosin light chain 1 [Source:HGNC Symbol;Acc:HGNC:7582] | 4105 | 0.340 | 0.5514 | No |
| 16 | VKORC1L1 | vitamin K epoxide reductase complex subunit 1 like 1 [Source:HGNC Symbol;Acc:HGNC:21492] | 4873 | 0.211 | 0.5359 | No |
| 17 | ESYT3 | extended synaptotagmin 3 [Source:HGNC Symbol;Acc:HGNC:24295] | 5158 | 0.168 | 0.5323 | No |
| 18 | LSM14A | LSM14A mRNA processing body assembly factor [Source:HGNC Symbol;Acc:HGNC:24489] | 6012 | 0.052 | 0.5106 | No |
| 19 | CPS1 | carbamoyl-phosphate synthase 1 [Source:HGNC Symbol;Acc:HGNC:2323] | 6088 | 0.040 | 0.5095 | No |
| 20 | PNPLA1 | patatin like phospholipase domain containing 1 [Source:HGNC Symbol;Acc:HGNC:21246] | 8168 | 0.000 | 0.4534 | No |
| 21 | KCNJ3 | potassium inwardly rectifying channel subfamily J member 3 [Source:HGNC Symbol;Acc:HGNC:6264] | 9833 | 0.000 | 0.4085 | No |
| 22 | MBOAT2 | membrane bound O-acyltransferase domain containing 2 [Source:HGNC Symbol;Acc:HGNC:25193] | 11444 | 0.000 | 0.3650 | No |
| 23 | PROZ | protein Z, vitamin K dependent plasma glycoprotein [Source:HGNC Symbol;Acc:HGNC:9460] | 11547 | 0.000 | 0.3623 | No |
| 24 | GLRA2 | glycine receptor alpha 2 [Source:HGNC Symbol;Acc:HGNC:4327] | 12905 | 0.000 | 0.3256 | No |
| 25 | RGS7 | regulator of G protein signaling 7 [Source:HGNC Symbol;Acc:HGNC:10003] | 14662 | 0.000 | 0.2782 | No |
| 26 | MDH1B | malate dehydrogenase 1B [Source:HGNC Symbol;Acc:HGNC:17836] | 16289 | 0.000 | 0.2343 | No |
| 27 | L3MBTL1 | L3MBTL histone methyl-lysine binding protein 1 [Source:HGNC Symbol;Acc:HGNC:15905] | 17515 | 0.000 | 0.2013 | No |
| 28 | ZNF594 | zinc finger protein 594 [Source:HGNC Symbol;Acc:HGNC:29392] | 21540 | 0.000 | 0.0926 | No |
| 29 | MYOCD | myocardin [Source:HGNC Symbol;Acc:HGNC:16067] | 24093 | 0.000 | 0.0237 | No |
| 30 | SLC4A5 | solute carrier family 4 member 5 [Source:HGNC Symbol;Acc:HGNC:18168] | 24985 | 0.000 | -0.0003 | No |
| 31 | OCLN | occludin [Source:HGNC Symbol;Acc:HGNC:8104] | 25805 | 0.000 | -0.0224 | No |
| 32 | SPZ1 | spermatogenic leucine zipper 1 [Source:HGNC Symbol;Acc:HGNC:30721] | 26100 | 0.000 | -0.0304 | No |
| 33 | FMO2 | flavin containing dimethylaniline monoxygenase 2 [Source:HGNC Symbol;Acc:HGNC:3770] | 26776 | 0.000 | -0.0486 | No |
| 34 | ZNF25 | zinc finger protein 25 [Source:HGNC Symbol;Acc:HGNC:13043] | 27186 | -0.012 | -0.0593 | No |
| 35 | PDZD2 | PDZ domain containing 2 [Source:HGNC Symbol;Acc:HGNC:18486] | 27412 | -0.050 | -0.0642 | No |
| 36 | NIPBL | NIPBL cohesin loading factor [Source:HGNC Symbol;Acc:HGNC:28862] | 27490 | -0.058 | -0.0649 | No |
| 37 | SV2C | synaptic vesicle glycoprotein 2C [Source:HGNC Symbol;Acc:HGNC:30670] | 27681 | -0.079 | -0.0681 | No |
| 38 | PITPNB | phosphatidylinositol transfer protein beta [Source:HGNC Symbol;Acc:HGNC:9002] | 28916 | -0.242 | -0.0954 | No |
| 39 | C1orf50 | chromosome 1 open reading frame 50 [Source:HGNC Symbol;Acc:HGNC:28795] | 29834 | -0.374 | -0.1110 | No |
| 40 | ICOS | inducible T cell costimulator [Source:HGNC Symbol;Acc:HGNC:5351] | 31070 | -0.570 | -0.1304 | No |
| 41 | CLK4 | CDC like kinase 4 [Source:HGNC Symbol;Acc:HGNC:13659] | 31273 | -0.602 | -0.1211 | No |
| 42 | MAPK10 | mitogen-activated protein kinase 10 [Source:HGNC Symbol;Acc:HGNC:6872] | 31560 | -0.661 | -0.1126 | No |
| 43 | TK2 | thymidine kinase 2 [Source:HGNC Symbol;Acc:HGNC:11831] | 32809 | -0.965 | -0.1226 | No |
| 44 | NHLRC3 | NHL repeat containing 3 [Source:HGNC Symbol;Acc:HGNC:33751] | 33548 | -1.186 | -0.1134 | No |
| 45 | KAT6B | lysine acetyltransferase 6B [Source:HGNC Symbol;Acc:HGNC:17582] | 33676 | -1.229 | -0.0867 | No |
| 46 | MEAK7 | MTOR associated protein, eak-7 homolog [Source:HGNC Symbol;Acc:HGNC:29325] | 33817 | -1.275 | -0.0593 | No |
| 47 | MBNL1 | muscleblind like splicing regulator 1 [Source:HGNC Symbol;Acc:HGNC:6923] | 33912 | -1.311 | -0.0296 | No |
| 48 | OSGEPL1 | O-sialoglycoprotein endopeptidase like 1 [Source:HGNC Symbol;Acc:HGNC:23075] | 35290 | -1.943 | -0.0192 | No |
| 49 | PM20D2 | peptidase M20 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21408] | 36243 | -2.762 | 0.0229 | No |