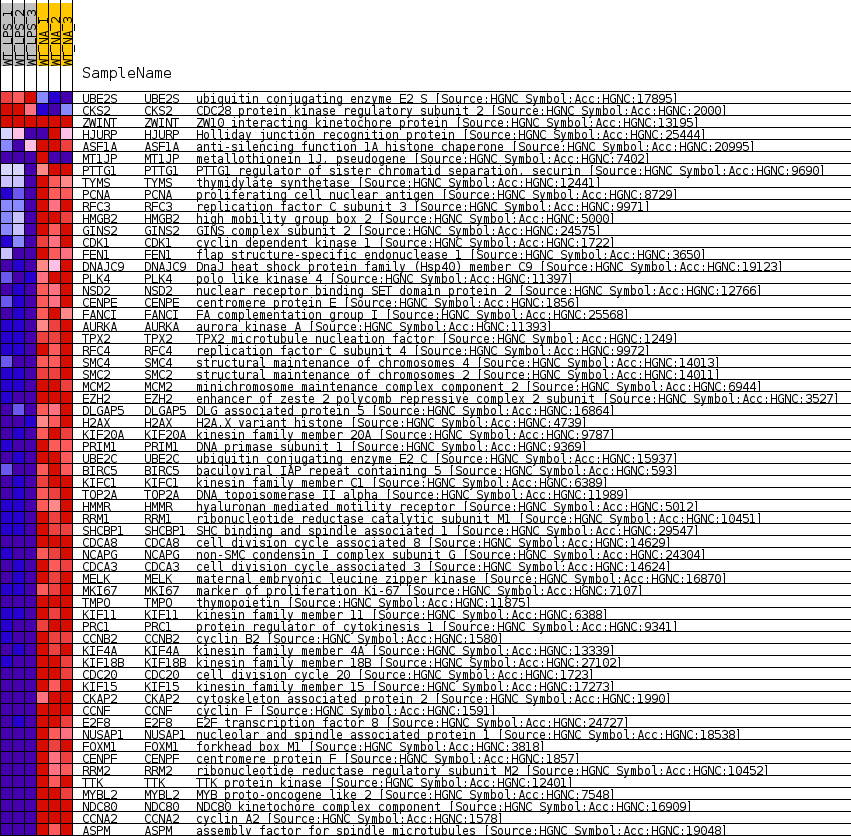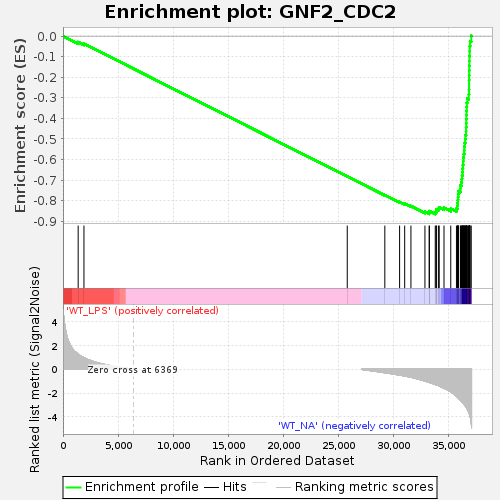
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_LPS_versus_WT_NA.greenbergcls.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | GNF2_CDC2 |
| Enrichment Score (ES) | -0.8655345 |
| Normalized Enrichment Score (NES) | -1.715905 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 1377 | 1.261 | -0.0292 | No |
| 2 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1902 | 0.962 | -0.0372 | No |
| 3 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 25804 | 0.000 | -0.6827 | No |
| 4 | HJURP | Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] | 29215 | -0.283 | -0.7730 | No |
| 5 | ASF1A | anti-silencing function 1A histone chaperone [Source:HGNC Symbol;Acc:HGNC:20995] | 30556 | -0.480 | -0.8061 | No |
| 6 | MT1JP | metallothionein 1J, pseudogene [Source:HGNC Symbol;Acc:HGNC:7402] | 31017 | -0.559 | -0.8150 | No |
| 7 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 31585 | -0.666 | -0.8261 | No |
| 8 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32856 | -0.976 | -0.8542 | No |
| 9 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33245 | -1.087 | -0.8577 | Yes |
| 10 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33258 | -1.093 | -0.8511 | Yes |
| 11 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33793 | -1.266 | -0.8575 | Yes |
| 12 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 33884 | -1.299 | -0.8517 | Yes |
| 13 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 33887 | -1.300 | -0.8435 | Yes |
| 14 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 34079 | -1.367 | -0.8399 | Yes |
| 15 | DNAJC9 | DnaJ heat shock protein family (Hsp40) member C9 [Source:HGNC Symbol;Acc:HGNC:19123] | 34151 | -1.393 | -0.8330 | Yes |
| 16 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 34588 | -1.576 | -0.8348 | Yes |
| 17 | NSD2 | nuclear receptor binding SET domain protein 2 [Source:HGNC Symbol;Acc:HGNC:12766] | 35201 | -1.889 | -0.8393 | Yes |
| 18 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 35717 | -2.250 | -0.8389 | Yes |
| 19 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35799 | -2.315 | -0.8264 | Yes |
| 20 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35816 | -2.333 | -0.8120 | Yes |
| 21 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 35837 | -2.347 | -0.7976 | Yes |
| 22 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35861 | -2.370 | -0.7832 | Yes |
| 23 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 35874 | -2.384 | -0.7684 | Yes |
| 24 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35901 | -2.411 | -0.7538 | Yes |
| 25 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 36076 | -2.578 | -0.7421 | Yes |
| 26 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 36079 | -2.583 | -0.7257 | Yes |
| 27 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 36174 | -2.692 | -0.7112 | Yes |
| 28 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36206 | -2.726 | -0.6947 | Yes |
| 29 | KIF20A | kinesin family member 20A [Source:HGNC Symbol;Acc:HGNC:9787] | 36227 | -2.746 | -0.6778 | Yes |
| 30 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 36266 | -2.780 | -0.6612 | Yes |
| 31 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 36279 | -2.795 | -0.6437 | Yes |
| 32 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 36304 | -2.820 | -0.6265 | Yes |
| 33 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 36332 | -2.865 | -0.6090 | Yes |
| 34 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 36339 | -2.870 | -0.5909 | Yes |
| 35 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 36383 | -2.908 | -0.5736 | Yes |
| 36 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36425 | -2.957 | -0.5559 | Yes |
| 37 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 36430 | -2.964 | -0.5372 | Yes |
| 38 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 36469 | -2.992 | -0.5192 | Yes |
| 39 | NCAPG | non-SMC condensin I complex subunit G [Source:HGNC Symbol;Acc:HGNC:24304] | 36516 | -3.057 | -0.5011 | Yes |
| 40 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36535 | -3.075 | -0.4820 | Yes |
| 41 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36586 | -3.138 | -0.4634 | Yes |
| 42 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36592 | -3.140 | -0.4436 | Yes |
| 43 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36607 | -3.171 | -0.4239 | Yes |
| 44 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 36611 | -3.176 | -0.4038 | Yes |
| 45 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36622 | -3.191 | -0.3838 | Yes |
| 46 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36630 | -3.207 | -0.3636 | Yes |
| 47 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 36650 | -3.229 | -0.3436 | Yes |
| 48 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 36663 | -3.250 | -0.3233 | Yes |
| 49 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 36709 | -3.356 | -0.3032 | Yes |
| 50 | KIF15 | kinesin family member 15 [Source:HGNC Symbol;Acc:HGNC:17273] | 36837 | -3.615 | -0.2836 | Yes |
| 51 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 36855 | -3.645 | -0.2609 | Yes |
| 52 | CCNF | cyclin F [Source:HGNC Symbol;Acc:HGNC:1591] | 36862 | -3.659 | -0.2379 | Yes |
| 53 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36863 | -3.662 | -0.2146 | Yes |
| 54 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36870 | -3.679 | -0.1914 | Yes |
| 55 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36876 | -3.698 | -0.1680 | Yes |
| 56 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36882 | -3.715 | -0.1446 | Yes |
| 57 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36891 | -3.747 | -0.1210 | Yes |
| 58 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 36894 | -3.758 | -0.0972 | Yes |
| 59 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36917 | -3.817 | -0.0735 | Yes |
| 60 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36922 | -3.842 | -0.0492 | Yes |
| 61 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36955 | -3.967 | -0.0249 | Yes |
| 62 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 37059 | -4.485 | 0.0008 | Yes |