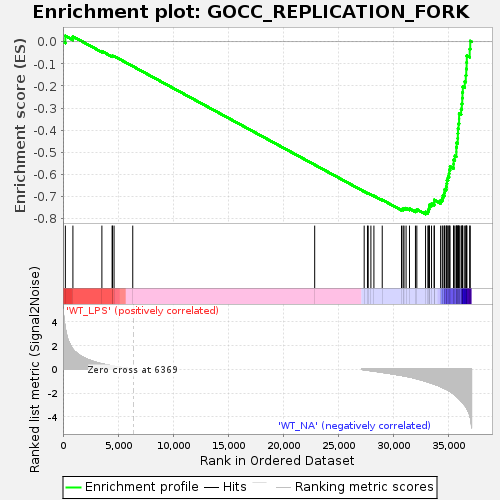
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.greenbergcls.cls #WT_LPS_versus_WT_NA.greenbergcls.cls #WT_LPS_versus_WT_NA_repos |
| Phenotype | greenbergcls.cls#WT_LPS_versus_WT_NA_repos |
| Upregulated in class | WT_NA |
| GeneSet | GOCC_REPLICATION_FORK |
| Enrichment Score (ES) | -0.77980816 |
| Normalized Enrichment Score (NES) | -1.5657734 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08942162 |
| FWER p-Value | 0.807 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | TREX1 | three prime repair exonuclease 1 [Source:HGNC Symbol;Acc:HGNC:12269] | 214 | 3.397 | 0.0266 | No |
| 2 | BCL6 | BCL6 transcription repressor [Source:HGNC Symbol;Acc:HGNC:1001] | 899 | 1.665 | 0.0240 | No |
| 3 | RTF2 | replication termination factor 2 [Source:HGNC Symbol;Acc:HGNC:15890] | 3530 | 0.453 | -0.0427 | No |
| 4 | SMARCAD1 | SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 [Source:HGNC Symbol;Acc:HGNC:18398] | 4461 | 0.276 | -0.0652 | No |
| 5 | TP53BP1 | tumor protein p53 binding protein 1 [Source:HGNC Symbol;Acc:HGNC:11999] | 4491 | 0.270 | -0.0634 | No |
| 6 | PLRG1 | pleiotropic regulator 1 [Source:HGNC Symbol;Acc:HGNC:9089] | 4635 | 0.247 | -0.0650 | No |
| 7 | UBE2B | ubiquitin conjugating enzyme E2 B [Source:HGNC Symbol;Acc:HGNC:12473] | 6332 | 0.005 | -0.1107 | No |
| 8 | RPA4 | replication protein A4 [Source:HGNC Symbol;Acc:HGNC:30305] | 22848 | 0.000 | -0.5567 | No |
| 9 | POLA2 | DNA polymerase alpha 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:30073] | 27341 | -0.041 | -0.6777 | No |
| 10 | SMARCA5 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 [Source:HGNC Symbol;Acc:HGNC:11101] | 27640 | -0.073 | -0.6850 | No |
| 11 | TP53 | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 27705 | -0.083 | -0.6860 | No |
| 12 | POLD4 | DNA polymerase delta 4, accessory subunit [Source:HGNC Symbol;Acc:HGNC:14106] | 27951 | -0.116 | -0.6915 | No |
| 13 | TIMELESS | timeless circadian regulator [Source:HGNC Symbol;Acc:HGNC:11813] | 28224 | -0.148 | -0.6974 | No |
| 14 | DONSON | DNA replication fork stabilization factor DONSON [Source:HGNC Symbol;Acc:HGNC:2993] | 28980 | -0.251 | -0.7154 | No |
| 15 | PRPF19 | pre-mRNA processing factor 19 [Source:HGNC Symbol;Acc:HGNC:17896] | 30728 | -0.511 | -0.7577 | No |
| 16 | NBN | nibrin [Source:HGNC Symbol;Acc:HGNC:7652] | 30870 | -0.536 | -0.7564 | No |
| 17 | DMAP1 | DNA methyltransferase 1 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18291] | 30955 | -0.549 | -0.7535 | No |
| 18 | CDC5L | cell division cycle 5 like [Source:HGNC Symbol;Acc:HGNC:1743] | 31145 | -0.581 | -0.7530 | No |
| 19 | ERCC5 | ERCC excision repair 5, endonuclease [Source:HGNC Symbol;Acc:HGNC:3437] | 31454 | -0.638 | -0.7553 | No |
| 20 | RAD18 | RAD18 E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:HGNC:18278] | 31995 | -0.762 | -0.7626 | No |
| 21 | XRCC2 | X-ray repair cross complementing 2 [Source:HGNC Symbol;Acc:HGNC:12829] | 32121 | -0.790 | -0.7584 | No |
| 22 | MCM10 | minichromosome maintenance 10 replication initiation factor [Source:HGNC Symbol;Acc:HGNC:18043] | 32913 | -0.991 | -0.7704 | Yes |
| 23 | BAZ1B | bromodomain adjacent to zinc finger domain 1B [Source:HGNC Symbol;Acc:HGNC:961] | 33124 | -1.054 | -0.7660 | Yes |
| 24 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 33196 | -1.074 | -0.7577 | Yes |
| 25 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33245 | -1.087 | -0.7486 | Yes |
| 26 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33258 | -1.093 | -0.7385 | Yes |
| 27 | SMARCAL1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a like 1 [Source:HGNC Symbol;Acc:HGNC:11102] | 33455 | -1.155 | -0.7328 | Yes |
| 28 | XRCC3 | X-ray repair cross complementing 3 [Source:HGNC Symbol;Acc:HGNC:12830] | 33700 | -1.236 | -0.7276 | Yes |
| 29 | XPA | XPA, DNA damage recognition and repair factor [Source:HGNC Symbol;Acc:HGNC:12814] | 33703 | -1.236 | -0.7159 | Yes |
| 30 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34278 | -1.445 | -0.7177 | Yes |
| 31 | RAD51D | RAD51 paralog D [Source:HGNC Symbol;Acc:HGNC:9823] | 34425 | -1.512 | -0.7072 | Yes |
| 32 | TONSL | tonsoku like, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:7801] | 34494 | -1.537 | -0.6944 | Yes |
| 33 | BCAS2 | BCAS2 pre-mRNA processing factor [Source:HGNC Symbol;Acc:HGNC:975] | 34631 | -1.596 | -0.6829 | Yes |
| 34 | WRN | WRN RecQ like helicase [Source:HGNC Symbol;Acc:HGNC:12791] | 34648 | -1.604 | -0.6680 | Yes |
| 35 | PRIM2 | DNA primase subunit 2 [Source:HGNC Symbol;Acc:HGNC:9370] | 34784 | -1.663 | -0.6558 | Yes |
| 36 | MRE11 | MRE11 homolog, double strand break repair nuclease [Source:HGNC Symbol;Acc:HGNC:7230] | 34836 | -1.689 | -0.6411 | Yes |
| 37 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 34857 | -1.696 | -0.6255 | Yes |
| 38 | RPA2 | replication protein A2 [Source:HGNC Symbol;Acc:HGNC:10290] | 34954 | -1.749 | -0.6114 | Yes |
| 39 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 35043 | -1.792 | -0.5967 | Yes |
| 40 | TIPIN | TIMELESS interacting protein [Source:HGNC Symbol;Acc:HGNC:30750] | 35066 | -1.805 | -0.5801 | Yes |
| 41 | TEX264 | testis expressed 264, ER-phagy receptor [Source:HGNC Symbol;Acc:HGNC:30247] | 35136 | -1.841 | -0.5644 | Yes |
| 42 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 35456 | -2.055 | -0.5535 | Yes |
| 43 | MMS22L | MMS22 like, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:21475] | 35460 | -2.057 | -0.5340 | Yes |
| 44 | POLA1 | DNA polymerase alpha 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9173] | 35571 | -2.133 | -0.5166 | Yes |
| 45 | POLD2 | DNA polymerase delta 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9176] | 35708 | -2.242 | -0.4989 | Yes |
| 46 | RADX | RPA1 related single stranded DNA binding protein, X-linked [Source:HGNC Symbol;Acc:HGNC:25486] | 35709 | -2.242 | -0.4776 | Yes |
| 47 | HMCES | 5-hydroxymethylcytosine binding, ES cell specific [Source:HGNC Symbol;Acc:HGNC:24446] | 35732 | -2.261 | -0.4566 | Yes |
| 48 | POLH | DNA polymerase eta [Source:HGNC Symbol;Acc:HGNC:9181] | 35834 | -2.346 | -0.4370 | Yes |
| 49 | ETAA1 | ETAA1 activator of ATR kinase [Source:HGNC Symbol;Acc:HGNC:24648] | 35844 | -2.356 | -0.4148 | Yes |
| 50 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35861 | -2.370 | -0.3926 | Yes |
| 51 | ZMIZ2 | zinc finger MIZ-type containing 2 [Source:HGNC Symbol;Acc:HGNC:22229] | 35907 | -2.416 | -0.3708 | Yes |
| 52 | RFC1 | replication factor C subunit 1 [Source:HGNC Symbol;Acc:HGNC:9969] | 35957 | -2.463 | -0.3487 | Yes |
| 53 | RAD51B | RAD51 paralog B [Source:HGNC Symbol;Acc:HGNC:9822] | 35959 | -2.468 | -0.3252 | Yes |
| 54 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36139 | -2.657 | -0.3047 | Yes |
| 55 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36206 | -2.726 | -0.2805 | Yes |
| 56 | PRIMPOL | primase and DNA directed polymerase [Source:HGNC Symbol;Acc:HGNC:26575] | 36244 | -2.762 | -0.2552 | Yes |
| 57 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 36266 | -2.780 | -0.2293 | Yes |
| 58 | UHRF1 | ubiquitin like with PHD and ring finger domains 1 [Source:HGNC Symbol;Acc:HGNC:12556] | 36296 | -2.816 | -0.2033 | Yes |
| 59 | BLM | BLM RecQ like helicase [Source:HGNC Symbol;Acc:HGNC:1058] | 36473 | -2.997 | -0.1794 | Yes |
| 60 | RFC2 | replication factor C subunit 2 [Source:HGNC Symbol;Acc:HGNC:9970] | 36566 | -3.117 | -0.1522 | Yes |
| 61 | RPA1 | replication protein A1 [Source:HGNC Symbol;Acc:HGNC:10289] | 36606 | -3.169 | -0.1231 | Yes |
| 62 | ZRANB3 | zinc finger RANBP2-type containing 3 [Source:HGNC Symbol;Acc:HGNC:25249] | 36641 | -3.218 | -0.0933 | Yes |
| 63 | HELB | DNA helicase B [Source:HGNC Symbol;Acc:HGNC:17196] | 36658 | -3.240 | -0.0629 | Yes |
| 64 | PIF1 | PIF1 5'-to-3' DNA helicase [Source:HGNC Symbol;Acc:HGNC:26220] | 36919 | -3.828 | -0.0335 | Yes |
| 65 | WDHD1 | WD repeat and HMG-box DNA binding protein 1 [Source:HGNC Symbol;Acc:HGNC:23170] | 36968 | -3.994 | 0.0033 | Yes |