Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
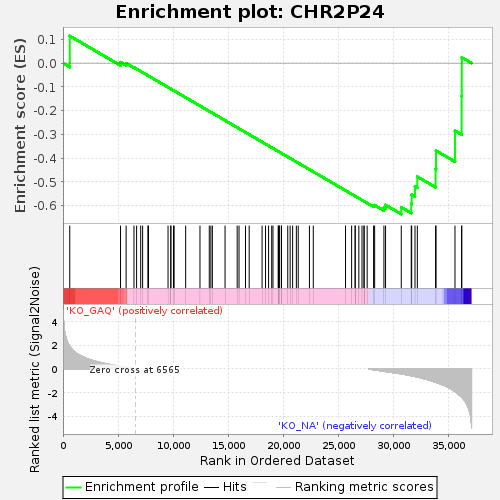
| Upregulated in class | KO_NA |
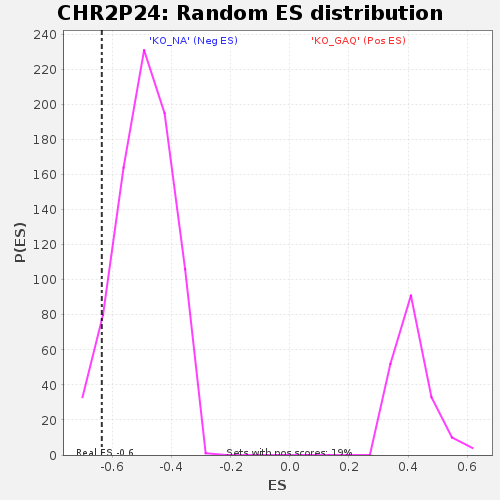
| GeneSet | CHR2P24 |
| Enrichment Score (ES) | -0.6349546 |
| Normalized Enrichment Score (NES) | -1.2860645 |
| Nominal p-value | 0.074074075 |
| FDR q-value | 0.5382412 |
| FWER p-Value | 1.0 |

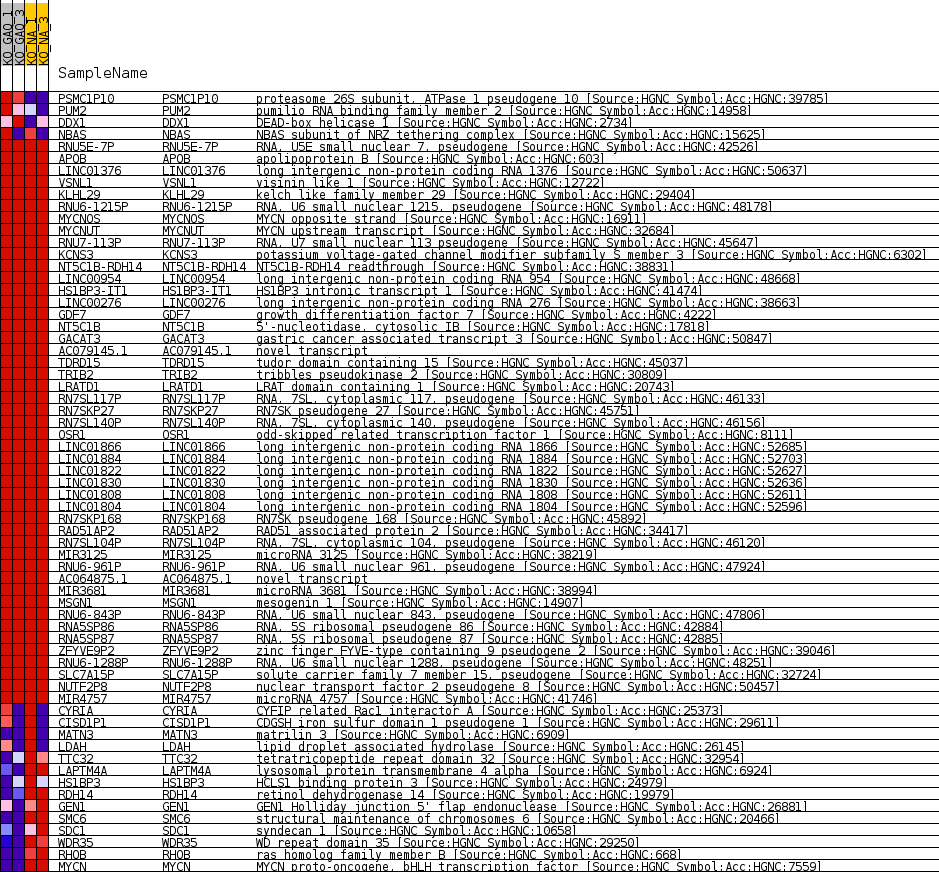
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PSMC1P10 | proteasome 26S subunit, ATPase 1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:39785] | 613 | 1.900 | 0.1143 | No |
| 2 | PUM2 | pumilio RNA binding family member 2 [Source:HGNC Symbol;Acc:HGNC:14958] | 5223 | 0.204 | 0.0039 | No |
| 3 | DDX1 | DEAD-box helicase 1 [Source:HGNC Symbol;Acc:HGNC:2734] | 5730 | 0.133 | -0.0006 | No |
| 4 | NBAS | NBAS subunit of NRZ tethering complex [Source:HGNC Symbol;Acc:HGNC:15625] | 6443 | 0.024 | -0.0182 | No |
| 5 | RNU5E-7P | RNA, U5E small nuclear 7, pseudogene [Source:HGNC Symbol;Acc:HGNC:42526] | 6683 | 0.000 | -0.0247 | No |
| 6 | APOB | apolipoprotein B [Source:HGNC Symbol;Acc:HGNC:603] | 7052 | 0.000 | -0.0346 | No |
| 7 | LINC01376 | long intergenic non-protein coding RNA 1376 [Source:HGNC Symbol;Acc:HGNC:50637] | 7228 | 0.000 | -0.0393 | No |
| 8 | VSNL1 | visinin like 1 [Source:HGNC Symbol;Acc:HGNC:12722] | 7706 | 0.000 | -0.0522 | No |
| 9 | KLHL29 | kelch like family member 29 [Source:HGNC Symbol;Acc:HGNC:29404] | 7762 | 0.000 | -0.0537 | No |
| 10 | RNU6-1215P | RNA, U6 small nuclear 1215, pseudogene [Source:HGNC Symbol;Acc:HGNC:48178] | 9536 | 0.000 | -0.1016 | No |
| 11 | MYCNOS | MYCN opposite strand [Source:HGNC Symbol;Acc:HGNC:16911] | 9784 | 0.000 | -0.1083 | No |
| 12 | MYCNUT | MYCN upstream transcript [Source:HGNC Symbol;Acc:HGNC:32684] | 9794 | 0.000 | -0.1085 | No |
| 13 | RNU7-113P | RNA, U7 small nuclear 113 pseudogene [Source:HGNC Symbol;Acc:HGNC:45647] | 10029 | 0.000 | -0.1148 | No |
| 14 | KCNS3 | potassium voltage-gated channel modifier subfamily S member 3 [Source:HGNC Symbol;Acc:HGNC:6302] | 10087 | 0.000 | -0.1164 | No |
| 15 | NT5C1B-RDH14 | NT5C1B-RDH14 readthrough [Source:HGNC Symbol;Acc:HGNC:38831] | 11135 | 0.000 | -0.1446 | No |
| 16 | LINC00954 | long intergenic non-protein coding RNA 954 [Source:HGNC Symbol;Acc:HGNC:48668] | 12434 | 0.000 | -0.1797 | No |
| 17 | HS1BP3-IT1 | HS1BP3 intronic transcript 1 [Source:HGNC Symbol;Acc:HGNC:41474] | 13279 | 0.000 | -0.2025 | No |
| 18 | LINC00276 | long intergenic non-protein coding RNA 276 [Source:HGNC Symbol;Acc:HGNC:38663] | 13424 | 0.000 | -0.2064 | No |
| 19 | GDF7 | growth differentiation factor 7 [Source:HGNC Symbol;Acc:HGNC:4222] | 13560 | 0.000 | -0.2100 | No |
| 20 | NT5C1B | 5'-nucleotidase, cytosolic IB [Source:HGNC Symbol;Acc:HGNC:17818] | 14711 | 0.000 | -0.2411 | No |
| 21 | GACAT3 | gastric cancer associated transcript 3 [Source:HGNC Symbol;Acc:HGNC:50847] | 15812 | 0.000 | -0.2708 | No |
| 22 | AC079145.1 | novel transcript | 15980 | 0.000 | -0.2753 | No |
| 23 | TDRD15 | tudor domain containing 15 [Source:HGNC Symbol;Acc:HGNC:45037] | 16568 | 0.000 | -0.2912 | No |
| 24 | TRIB2 | tribbles pseudokinase 2 [Source:HGNC Symbol;Acc:HGNC:30809] | 16902 | 0.000 | -0.3001 | No |
| 25 | LRATD1 | LRAT domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20743] | 18072 | 0.000 | -0.3317 | No |
| 26 | RN7SL117P | RNA, 7SL, cytoplasmic 117, pseudogene [Source:HGNC Symbol;Acc:HGNC:46133] | 18386 | 0.000 | -0.3402 | No |
| 27 | RN7SKP27 | RN7SK pseudogene 27 [Source:HGNC Symbol;Acc:HGNC:45751] | 18664 | 0.000 | -0.3477 | No |
| 28 | RN7SL140P | RNA, 7SL, cytoplasmic 140, pseudogene [Source:HGNC Symbol;Acc:HGNC:46156] | 18945 | 0.000 | -0.3552 | No |
| 29 | OSR1 | odd-skipped related transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:8111] | 19060 | 0.000 | -0.3583 | No |
| 30 | LINC01866 | long intergenic non-protein coding RNA 1866 [Source:HGNC Symbol;Acc:HGNC:52685] | 19533 | 0.000 | -0.3710 | No |
| 31 | LINC01884 | long intergenic non-protein coding RNA 1884 [Source:HGNC Symbol;Acc:HGNC:52703] | 19571 | 0.000 | -0.3720 | No |
| 32 | LINC01822 | long intergenic non-protein coding RNA 1822 [Source:HGNC Symbol;Acc:HGNC:52627] | 19607 | 0.000 | -0.3730 | No |
| 33 | LINC01830 | long intergenic non-protein coding RNA 1830 [Source:HGNC Symbol;Acc:HGNC:52636] | 19667 | 0.000 | -0.3746 | No |
| 34 | LINC01808 | long intergenic non-protein coding RNA 1808 [Source:HGNC Symbol;Acc:HGNC:52611] | 19834 | 0.000 | -0.3791 | No |
| 35 | LINC01804 | long intergenic non-protein coding RNA 1804 [Source:HGNC Symbol;Acc:HGNC:52596] | 19837 | 0.000 | -0.3791 | No |
| 36 | RN7SKP168 | RN7SK pseudogene 168 [Source:HGNC Symbol;Acc:HGNC:45892] | 20403 | 0.000 | -0.3944 | No |
| 37 | RAD51AP2 | RAD51 associated protein 2 [Source:HGNC Symbol;Acc:HGNC:34417] | 20620 | 0.000 | -0.4002 | No |
| 38 | RN7SL104P | RNA, 7SL, cytoplasmic 104, pseudogene [Source:HGNC Symbol;Acc:HGNC:46120] | 20846 | 0.000 | -0.4063 | No |
| 39 | MIR3125 | microRNA 3125 [Source:HGNC Symbol;Acc:HGNC:38219] | 21183 | 0.000 | -0.4154 | No |
| 40 | RNU6-961P | RNA, U6 small nuclear 961, pseudogene [Source:HGNC Symbol;Acc:HGNC:47924] | 21354 | 0.000 | -0.4200 | No |
| 41 | AC064875.1 | novel transcript | 22370 | 0.000 | -0.4474 | No |
| 42 | MIR3681 | microRNA 3681 [Source:HGNC Symbol;Acc:HGNC:38994] | 22720 | 0.000 | -0.4568 | No |
| 43 | MSGN1 | mesogenin 1 [Source:HGNC Symbol;Acc:HGNC:14907] | 25648 | 0.000 | -0.5358 | No |
| 44 | RNU6-843P | RNA, U6 small nuclear 843, pseudogene [Source:HGNC Symbol;Acc:HGNC:47806] | 26201 | 0.000 | -0.5508 | No |
| 45 | RNA5SP86 | RNA, 5S ribosomal pseudogene 86 [Source:HGNC Symbol;Acc:HGNC:42884] | 26522 | 0.000 | -0.5594 | No |
| 46 | RNA5SP87 | RNA, 5S ribosomal pseudogene 87 [Source:HGNC Symbol;Acc:HGNC:42885] | 26526 | 0.000 | -0.5595 | No |
| 47 | ZFYVE9P2 | zinc finger FYVE-type containing 9 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:39046] | 26856 | 0.000 | -0.5684 | No |
| 48 | RNU6-1288P | RNA, U6 small nuclear 1288, pseudogene [Source:HGNC Symbol;Acc:HGNC:48251] | 27143 | 0.000 | -0.5761 | No |
| 49 | SLC7A15P | solute carrier family 7 member 15, pseudogene [Source:HGNC Symbol;Acc:HGNC:32724] | 27330 | 0.000 | -0.5811 | No |
| 50 | NUTF2P8 | nuclear transport factor 2 pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:50457] | 27332 | 0.000 | -0.5811 | No |
| 51 | MIR4757 | microRNA 4757 [Source:HGNC Symbol;Acc:HGNC:41746] | 27611 | 0.000 | -0.5886 | No |
| 52 | CYRIA | CYFIP related Rac1 interactor A [Source:HGNC Symbol;Acc:HGNC:25373] | 28194 | -0.063 | -0.6000 | No |
| 53 | CISD1P1 | CDGSH iron sulfur domain 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:29611] | 28291 | -0.076 | -0.5974 | No |
| 54 | MATN3 | matrilin 3 [Source:HGNC Symbol;Acc:HGNC:6909] | 29133 | -0.188 | -0.6071 | Yes |
| 55 | LDAH | lipid droplet associated hydrolase [Source:HGNC Symbol;Acc:HGNC:26145] | 29282 | -0.213 | -0.5965 | Yes |
| 56 | TTC32 | tetratricopeptide repeat domain 32 [Source:HGNC Symbol;Acc:HGNC:32954] | 30708 | -0.418 | -0.6062 | Yes |
| 57 | LAPTM4A | lysosomal protein transmembrane 4 alpha [Source:HGNC Symbol;Acc:HGNC:6924] | 31613 | -0.563 | -0.5918 | Yes |
| 58 | HS1BP3 | HCLS1 binding protein 3 [Source:HGNC Symbol;Acc:HGNC:24979] | 31654 | -0.570 | -0.5537 | Yes |
| 59 | RDH14 | retinol dehydrogenase 14 [Source:HGNC Symbol;Acc:HGNC:19979] | 31955 | -0.630 | -0.5184 | Yes |
| 60 | GEN1 | GEN1 Holliday junction 5' flap endonuclease [Source:HGNC Symbol;Acc:HGNC:26881] | 32166 | -0.675 | -0.4776 | Yes |
| 61 | SMC6 | structural maintenance of chromosomes 6 [Source:HGNC Symbol;Acc:HGNC:20466] | 33822 | -1.119 | -0.4452 | Yes |
| 62 | SDC1 | syndecan 1 [Source:HGNC Symbol;Acc:HGNC:10658] | 33863 | -1.134 | -0.3682 | Yes |
| 63 | WDR35 | WD repeat domain 35 [Source:HGNC Symbol;Acc:HGNC:29250] | 35584 | -1.893 | -0.2843 | Yes |
| 64 | RHOB | ras homolog family member B [Source:HGNC Symbol;Acc:HGNC:668] | 36189 | -2.355 | -0.1384 | Yes |
| 65 | MYCN | MYCN proto-oncogene, bHLH transcription factor [Source:HGNC Symbol;Acc:HGNC:7559] | 36198 | -2.363 | 0.0241 | Yes |