Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR6P25 |
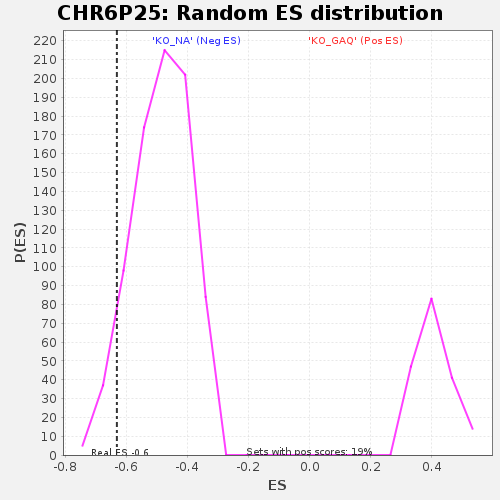
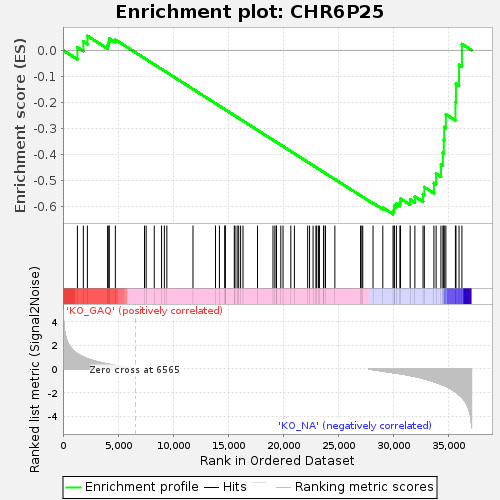
| Enrichment Score (ES) | -0.63079095 |
| Normalized Enrichment Score (NES) | -1.2996986 |
| Nominal p-value | 0.057668712 |
| FDR q-value | 0.54903 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PSMC1P11 | proteasome 26S subunit, ATPase 1 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:39786] | 1309 | 1.267 | 0.0112 | No |
| 2 | FAM136BP | family with sequence similarity 136 member B, pseudogene [Source:HGNC Symbol;Acc:HGNC:21110] | 1847 | 0.999 | 0.0334 | No |
| 3 | AL033523.1 | novel transcript | 2214 | 0.857 | 0.0550 | No |
| 4 | IRF4 | interferon regulatory factor 4 [Source:HGNC Symbol;Acc:HGNC:6119] | 4048 | 0.407 | 0.0205 | No |
| 5 | HNRNPA1P37 | heterogeneous nuclear ribonucleoprotein A1 pseudogene 37 [Source:HGNC Symbol;Acc:HGNC:48766] | 4159 | 0.390 | 0.0318 | No |
| 6 | LYRM4 | LYR motif containing 4 [Source:HGNC Symbol;Acc:HGNC:21365] | 4195 | 0.380 | 0.0449 | No |
| 7 | PSMG4 | proteasome assembly chaperone 4 [Source:HGNC Symbol;Acc:HGNC:21108] | 4753 | 0.287 | 0.0404 | No |
| 8 | TDGF1P4 | teratocarcinoma-derived growth factor 1 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:11704] | 7399 | 0.000 | -0.0311 | No |
| 9 | LINC01600 | long intergenic non-protein coding RNA 1600 [Source:HGNC Symbol;Acc:HGNC:21600] | 7549 | 0.000 | -0.0351 | No |
| 10 | LINC01011 | long intergenic non-protein coding RNA 1011 [Source:HGNC Symbol;Acc:HGNC:33812] | 8284 | 0.000 | -0.0549 | No |
| 11 | AL159166.1 | novel transcript | 8947 | 0.000 | -0.0728 | No |
| 12 | C6orf201 | chromosome 6 open reading frame 201 [Source:HGNC Symbol;Acc:HGNC:21620] | 9200 | 0.000 | -0.0796 | No |
| 13 | LY86 | lymphocyte antigen 86 [Source:HGNC Symbol;Acc:HGNC:16837] | 9427 | 0.000 | -0.0857 | No |
| 14 | OR4F1P | olfactory receptor family 4 subfamily F member 1 pseudogene [Source:HGNC Symbol;Acc:HGNC:8298] | 11800 | 0.000 | -0.1498 | No |
| 15 | AL021328.1 | novel transcript, antisense to FARS2 | 13844 | 0.000 | -0.2050 | No |
| 16 | FAM217A | family with sequence similarity 217 member A [Source:HGNC Symbol;Acc:HGNC:21362] | 14202 | 0.000 | -0.2146 | No |
| 17 | LINC02533 | long intergenic non-protein coding RNA 2533 [Source:HGNC Symbol;Acc:HGNC:53566] | 14698 | 0.000 | -0.2280 | No |
| 18 | LINC02525 | long intergenic non-protein coding RNA 2525 [Source:HGNC Symbol;Acc:HGNC:53545] | 14713 | 0.000 | -0.2284 | No |
| 19 | LINC02521 | long intergenic non-protein coding RNA 2521 [Source:HGNC Symbol;Acc:HGNC:53560] | 14718 | 0.000 | -0.2285 | No |
| 20 | MIR5683 | microRNA 5683 [Source:HGNC Symbol;Acc:HGNC:43453] | 15549 | 0.000 | -0.2509 | No |
| 21 | AL035696.3 | novel transcript | 15614 | 0.000 | -0.2526 | No |
| 22 | RN7SL554P | RNA, 7SL, cytoplasmic 554, pseudogene [Source:HGNC Symbol;Acc:HGNC:46570] | 15806 | 0.000 | -0.2578 | No |
| 23 | HMGN2P28 | high mobility group nucleosomal binding domain 2 pseudogene 28 [Source:HGNC Symbol;Acc:HGNC:39395] | 15947 | 0.000 | -0.2616 | No |
| 24 | SNAPC5P1 | small nuclear RNA activating complex polypeptide 5 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:38667] | 16123 | 0.000 | -0.2663 | No |
| 25 | WBP1LP12 | WBP1L pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:51472] | 16340 | 0.000 | -0.2721 | No |
| 26 | PPP1R3G | protein phosphatase 1 regulatory subunit 3G [Source:HGNC Symbol;Acc:HGNC:14945] | 17656 | 0.000 | -0.3076 | No |
| 27 | LY86-AS1 | LY86 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:26593] | 19062 | 0.000 | -0.3456 | No |
| 28 | AL022097.1 | novel transcript, antisense to FARS2 | 19235 | 0.000 | -0.3502 | No |
| 29 | RNA5SP202 | RNA, 5S ribosomal pseudogene 202 [Source:HGNC Symbol;Acc:HGNC:43102] | 19372 | 0.000 | -0.3539 | No |
| 30 | RNA5SP201 | RNA, 5S ribosomal pseudogene 201 [Source:HGNC Symbol;Acc:HGNC:43101] | 19373 | 0.000 | -0.3539 | No |
| 31 | SERPINB9P1 | serpin family B member 9 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:28590] | 19772 | 0.000 | -0.3647 | No |
| 32 | RN7SL352P | RNA, 7SL, cytoplasmic 352, pseudogene [Source:HGNC Symbol;Acc:HGNC:46368] | 19974 | 0.000 | -0.3701 | No |
| 33 | SERPINB8P1 | serpin family B member 8 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:8953] | 20682 | 0.000 | -0.3892 | No |
| 34 | PXDC1 | PX domain containing 1 [Source:HGNC Symbol;Acc:HGNC:21361] | 21006 | 0.000 | -0.3979 | No |
| 35 | CNN3P1 | calponin 3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:38666] | 22209 | 0.000 | -0.4304 | No |
| 36 | GMDS-DT | GMDS divergent transcript [Source:HGNC Symbol;Acc:HGNC:48993] | 22383 | 0.000 | -0.4351 | No |
| 37 | MIR3691 | microRNA 3691 [Source:HGNC Symbol;Acc:HGNC:38909] | 22702 | 0.000 | -0.4437 | No |
| 38 | F13A1 | coagulation factor XIII A chain [Source:HGNC Symbol;Acc:HGNC:3531] | 22959 | 0.000 | -0.4506 | No |
| 39 | HUS1B | HUS1 checkpoint clamp component B [Source:HGNC Symbol;Acc:HGNC:16485] | 23031 | 0.000 | -0.4525 | No |
| 40 | CICP18 | capicua transcriptional repressor pseudogene 18 [Source:HGNC Symbol;Acc:HGNC:48828] | 23187 | 0.000 | -0.4567 | No |
| 41 | FAM50B | family with sequence similarity 50 member B [Source:HGNC Symbol;Acc:HGNC:18789] | 23280 | 0.000 | -0.4592 | No |
| 42 | MYLK4 | myosin light chain kinase family member 4 [Source:HGNC Symbol;Acc:HGNC:27972] | 23664 | 0.000 | -0.4695 | No |
| 43 | FOXQ1 | forkhead box Q1 [Source:HGNC Symbol;Acc:HGNC:20951] | 23665 | 0.000 | -0.4695 | No |
| 44 | FOXF2 | forkhead box F2 [Source:HGNC Symbol;Acc:HGNC:3810] | 23827 | 0.000 | -0.4739 | No |
| 45 | RN7SL221P | RNA, 7SL, cytoplasmic 221, pseudogene [Source:HGNC Symbol;Acc:HGNC:46237] | 24680 | 0.000 | -0.4969 | No |
| 46 | SLC22A23 | solute carrier family 22 member 23 [Source:HGNC Symbol;Acc:HGNC:21106] | 27011 | 0.000 | -0.5598 | No |
| 47 | FOXCUT | FOXC1 upstream transcript [Source:HGNC Symbol;Acc:HGNC:50650] | 27129 | 0.000 | -0.5630 | No |
| 48 | MIR4645 | microRNA 4645 [Source:HGNC Symbol;Acc:HGNC:41689] | 27212 | 0.000 | -0.5652 | No |
| 49 | PRPF4B | pre-mRNA processing factor 4B [Source:HGNC Symbol;Acc:HGNC:17346] | 28145 | -0.055 | -0.5883 | No |
| 50 | EXOC2 | exocyst complex component 2 [Source:HGNC Symbol;Acc:HGNC:24968] | 29032 | -0.177 | -0.6058 | No |
| 51 | RPP40 | ribonuclease P/MRP subunit p40 [Source:HGNC Symbol;Acc:HGNC:20992] | 29959 | -0.311 | -0.6194 | Yes |
| 52 | RIPK1 | receptor interacting serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:10019] | 30064 | -0.325 | -0.6102 | Yes |
| 53 | CDYL | chromodomain Y like [Source:HGNC Symbol;Acc:HGNC:1811] | 30079 | -0.327 | -0.5986 | Yes |
| 54 | WRNIP1 | WRN helicase interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:20876] | 30266 | -0.349 | -0.5908 | Yes |
| 55 | TUBB2BP1 | tubulin beta 2B class IIb pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:42332] | 30584 | -0.399 | -0.5847 | Yes |
| 56 | BTF3P7 | basic transcription factor 3 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:38569] | 30643 | -0.408 | -0.5713 | Yes |
| 57 | NQO2 | N-ribosyldihydronicotinamide:quinone reductase 2 [Source:HGNC Symbol;Acc:HGNC:7856] | 31516 | -0.548 | -0.5747 | Yes |
| 58 | GLRX3P2 | glutaredoxin 3 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:49788] | 31943 | -0.627 | -0.5632 | Yes |
| 59 | SERPINB1 | serpin family B member 1 [Source:HGNC Symbol;Acc:HGNC:3311] | 32683 | -0.791 | -0.5540 | Yes |
| 60 | TUBB2A | tubulin beta 2A class IIa [Source:HGNC Symbol;Acc:HGNC:12412] | 32801 | -0.823 | -0.5270 | Yes |
| 61 | FARS2 | phenylalanyl-tRNA synthetase 2, mitochondrial [Source:HGNC Symbol;Acc:HGNC:21062] | 33672 | -1.072 | -0.5111 | Yes |
| 62 | GMDS | GDP-mannose 4,6-dehydratase [Source:HGNC Symbol;Acc:HGNC:4369] | 33872 | -1.136 | -0.4747 | Yes |
| 63 | NRN1 | neuritin 1 [Source:HGNC Symbol;Acc:HGNC:17972] | 34308 | -1.283 | -0.4394 | Yes |
| 64 | PKMP5 | pyruvate kinase M1/2 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:44247] | 34489 | -1.349 | -0.3947 | Yes |
| 65 | FOXC1 | forkhead box C1 [Source:HGNC Symbol;Acc:HGNC:3800] | 34569 | -1.379 | -0.3461 | Yes |
| 66 | SERPINB9 | serpin family B member 9 [Source:HGNC Symbol;Acc:HGNC:8955] | 34604 | -1.393 | -0.2958 | Yes |
| 67 | SERPINB6 | serpin family B member 6 [Source:HGNC Symbol;Acc:HGNC:8950] | 34755 | -1.444 | -0.2468 | Yes |
| 68 | BPHL | biphenyl hydrolase like [Source:HGNC Symbol;Acc:HGNC:1094] | 35624 | -1.921 | -0.1997 | Yes |
| 69 | TUBB2B | tubulin beta 2B class IIb [Source:HGNC Symbol;Acc:HGNC:30829] | 35673 | -1.956 | -0.1291 | Yes |
| 70 | ECI2 | enoyl-CoA delta isomerase 2 [Source:HGNC Symbol;Acc:HGNC:14601] | 35952 | -2.174 | -0.0567 | Yes |
| 71 | DUSP22 | dual specificity phosphatase 22 [Source:HGNC Symbol;Acc:HGNC:16077] | 36214 | -2.380 | 0.0237 | Yes |