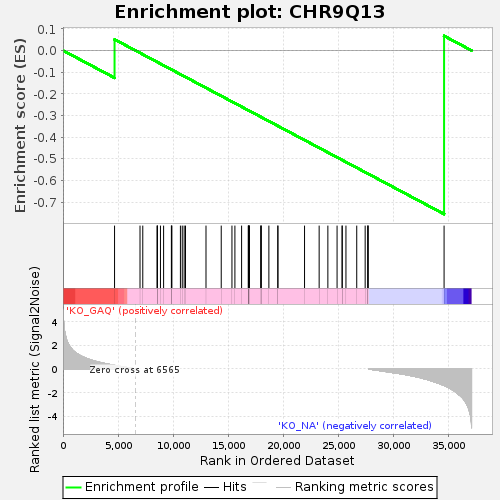
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR9Q13 |
| Enrichment Score (ES) | -0.755132 |
| Normalized Enrichment Score (NES) | -1.4489137 |
| Nominal p-value | 0.010376135 |
| FDR q-value | 0.50267285 |
| FWER p-Value | 0.685 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | BMS1P10 | BMS1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:49154] | 4680 | 0.300 | 0.0512 | No |
| 2 | LINC01410 | long intergenic non-protein coding RNA 1410 [Source:HGNC Symbol;Acc:HGNC:50702] | 6991 | 0.000 | -0.0112 | No |
| 3 | RAB28P2 | RAB28, member RAS oncogene family pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:50381] | 7242 | 0.000 | -0.0179 | No |
| 4 | AQP7P2 | aquaporin 7 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:32049] | 8557 | 0.000 | -0.0534 | No |
| 5 | AQP7P1 | aquaporin 7 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:32048] | 8559 | 0.000 | -0.0534 | No |
| 6 | AQP7P4 | aquaporin 7 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:32056] | 8562 | 0.000 | -0.0535 | No |
| 7 | LINC01189 | long intergenic non-protein coding RNA 1189 [Source:HGNC Symbol;Acc:HGNC:49592] | 8847 | 0.000 | -0.0612 | No |
| 8 | SNX18P9 | sorting nexin 18 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:39617] | 9133 | 0.000 | -0.0689 | No |
| 9 | AL591379.1 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like pseudogene [Source:NCBI gene (formerly Entrezgene);Acc:100996643] | 9838 | 0.000 | -0.0879 | No |
| 10 | CDK2AP2P3 | cyclin dependent kinase 2 associated protein 2 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:38494] | 9869 | 0.000 | -0.0887 | No |
| 11 | CDK2AP2P2 | cyclin dependent kinase 2 associated protein 2 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:38493] | 9870 | 0.000 | -0.0887 | No |
| 12 | ADGRF5P1 | adhesion G protein-coupled receptor F5 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:32922] | 10658 | 0.000 | -0.1099 | No |
| 13 | FAM27C | family with sequence similarity 27 member C [Source:HGNC Symbol;Acc:HGNC:23668] | 10825 | 0.000 | -0.1144 | No |
| 14 | GXYLT1P6 | GXYLT1 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:50425] | 10997 | 0.000 | -0.1190 | No |
| 15 | ANKRD20A4P | ankyrin repeat domain 20 family member A4, pseudogene [Source:HGNC Symbol;Acc:HGNC:31982] | 11120 | 0.000 | -0.1223 | No |
| 16 | RNU6-1193P | RNA, U6 small nuclear 1193, pseudogene [Source:HGNC Symbol;Acc:HGNC:48156] | 12981 | 0.000 | -0.1725 | No |
| 17 | ATP5F1AP7 | ATP synthase F1 subunit alpha pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:37665] | 14364 | 0.000 | -0.2098 | No |
| 18 | RBPJP7 | RBPJ pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:37487] | 15337 | 0.000 | -0.2360 | No |
| 19 | CNN2P3 | calponin 2 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:39528] | 15605 | 0.000 | -0.2433 | No |
| 20 | SNORA70 | small nucleolar RNA, H/ACA box 70 [Source:HGNC Symbol;Acc:HGNC:10231] | 16213 | 0.000 | -0.2596 | No |
| 21 | CNTNAP3P1 | CNTNAP3 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:49591] | 16826 | 0.000 | -0.2762 | No |
| 22 | CNTNAP3P5 | CNTNAP3 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:49588] | 16828 | 0.000 | -0.2762 | No |
| 23 | FGF7P6 | fibroblast growth factor 7 pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:27852] | 16879 | 0.000 | -0.2775 | No |
| 24 | FGF7P7 | fibroblast growth factor 7 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:33589] | 16880 | 0.000 | -0.2775 | No |
| 25 | FGF7P8 | fibroblast growth factor 7 pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:34516] | 16881 | 0.000 | -0.2775 | No |
| 26 | BMS1P9 | BMS1 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:49155] | 16919 | 0.000 | -0.2785 | No |
| 27 | FKBP4P7 | FKBP prolyl isomerase 4 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:50376] | 17955 | 0.000 | -0.3065 | No |
| 28 | AL512625.1 | uncharacterized LOC554249 [Source:NCBI gene (formerly Entrezgene);Acc:554249] | 17985 | 0.000 | -0.3073 | No |
| 29 | RN7SL544P | RNA, 7SL, cytoplasmic 544, pseudogene [Source:HGNC Symbol;Acc:HGNC:46560] | 17990 | 0.000 | -0.3074 | No |
| 30 | DUX4L50 | double homeobox 4 like 50 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:51788] | 18689 | 0.000 | -0.3262 | No |
| 31 | RNA5SP284 | RNA, 5S ribosomal pseudogene 284 [Source:HGNC Symbol;Acc:HGNC:43184] | 19505 | 0.000 | -0.3482 | No |
| 32 | RNA5SP283 | RNA, 5S ribosomal pseudogene 283 [Source:HGNC Symbol;Acc:HGNC:43183] | 19506 | 0.000 | -0.3482 | No |
| 33 | SDR42E1P3 | short chain dehydrogenase/reductase family 42E, member 1 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:51834] | 21927 | 0.000 | -0.4135 | No |
| 34 | CYP4F25P | cytochrome P450 family 4 subfamily F member 25, pseudogene [Source:HGNC Symbol;Acc:HGNC:39949] | 23253 | 0.000 | -0.4493 | No |
| 35 | BMS1P11 | BMS1 pseudogene 11 [Source:HGNC Symbol;Acc:HGNC:49156] | 24044 | 0.000 | -0.4706 | No |
| 36 | FRG1JP | FSHD region gene 1 family member J, pseudogene [Source:HGNC Symbol;Acc:HGNC:51768] | 24880 | 0.000 | -0.4932 | No |
| 37 | PTGER4P3 | prostaglandin E receptor 4 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:38498] | 25342 | 0.000 | -0.5056 | No |
| 38 | PTGER4P2 | prostaglandin E receptor 4 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:9598] | 25343 | 0.000 | -0.5056 | No |
| 39 | MIR4477B | microRNA 4477b [Source:HGNC Symbol;Acc:HGNC:41898] | 25686 | 0.000 | -0.5148 | No |
| 40 | IGKV1OR-2 | immunoglobulin kappa variable 1/OR-2 (pseudogene) [Source:HGNC Symbol;Acc:HGNC:5761] | 26665 | 0.000 | -0.5412 | No |
| 41 | RNU6-1293P | RNA, U6 small nuclear 1293, pseudogene [Source:HGNC Symbol;Acc:HGNC:48256] | 27430 | 0.000 | -0.5619 | No |
| 42 | MYO5BP3 | myosin VB pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:38497] | 27681 | 0.000 | -0.5686 | No |
| 43 | MYO5BP2 | myosin VB pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:38496] | 27682 | 0.000 | -0.5686 | No |
| 44 | RN7SL722P | RNA, 7SL, cytoplasmic 722, pseudogene [Source:HGNC Symbol;Acc:HGNC:46738] | 27685 | 0.000 | -0.5687 | Yes |
| 45 | BNIP3P4 | BCL2 interacting protein 3 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:39657] | 34594 | -1.389 | 0.0674 | Yes |

