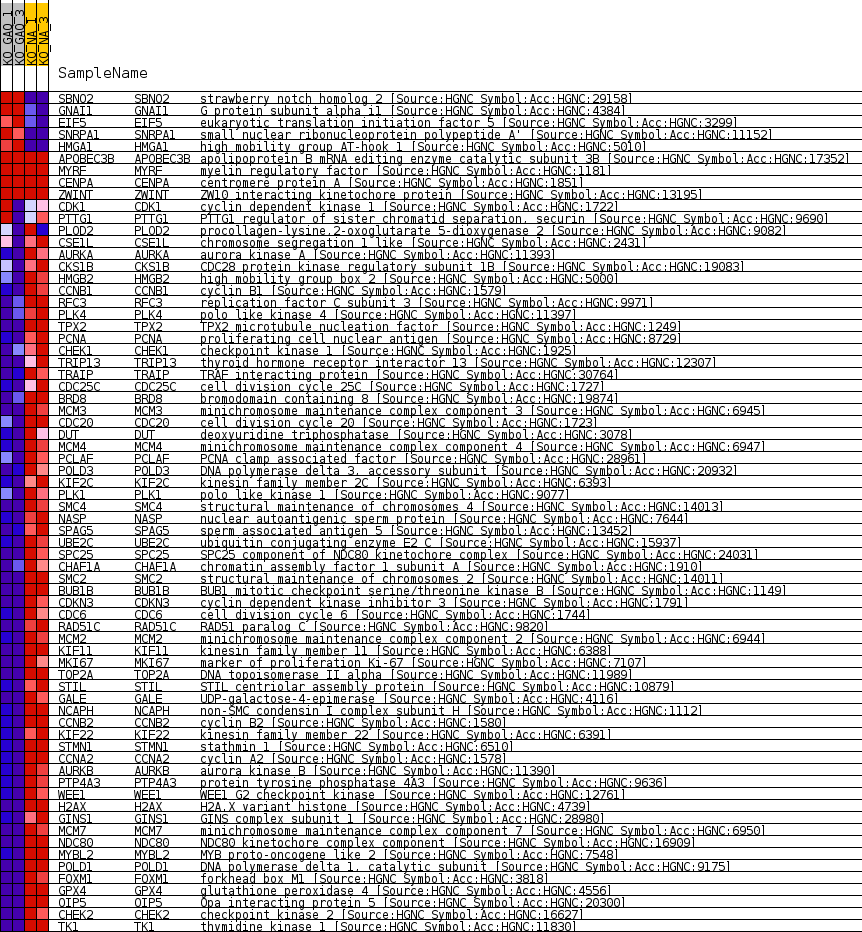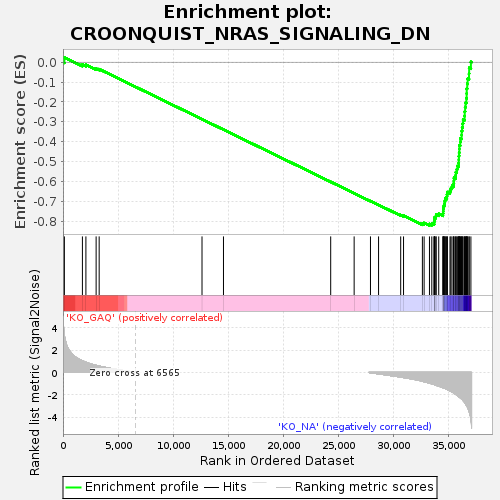
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CROONQUIST_NRAS_SIGNALING_DN |
| Enrichment Score (ES) | -0.8226935 |
| Normalized Enrichment Score (NES) | -1.674777 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0031401755 |
| FWER p-Value | 0.051 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SBNO2 | strawberry notch homolog 2 [Source:HGNC Symbol;Acc:HGNC:29158] | 126 | 3.278 | 0.0241 | No |
| 2 | GNAI1 | G protein subunit alpha i1 [Source:HGNC Symbol;Acc:HGNC:4384] | 1762 | 1.031 | -0.0114 | No |
| 3 | EIF5 | eukaryotic translation initiation factor 5 [Source:HGNC Symbol;Acc:HGNC:3299] | 2079 | 0.901 | -0.0124 | No |
| 4 | SNRPA1 | small nuclear ribonucleoprotein polypeptide A' [Source:HGNC Symbol;Acc:HGNC:11152] | 3007 | 0.629 | -0.0322 | No |
| 5 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 3283 | 0.564 | -0.0349 | No |
| 6 | APOBEC3B | apolipoprotein B mRNA editing enzyme catalytic subunit 3B [Source:HGNC Symbol;Acc:HGNC:17352] | 12619 | 0.000 | -0.2870 | No |
| 7 | MYRF | myelin regulatory factor [Source:HGNC Symbol;Acc:HGNC:1181] | 14568 | 0.000 | -0.3396 | No |
| 8 | CENPA | centromere protein A [Source:HGNC Symbol;Acc:HGNC:1851] | 24301 | 0.000 | -0.6025 | No |
| 9 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26430 | 0.000 | -0.6600 | No |
| 10 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27912 | -0.021 | -0.6998 | No |
| 11 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 28646 | -0.123 | -0.7186 | No |
| 12 | PLOD2 | procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 [Source:HGNC Symbol;Acc:HGNC:9082] | 30655 | -0.410 | -0.7694 | No |
| 13 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 30914 | -0.450 | -0.7726 | No |
| 14 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 32625 | -0.776 | -0.8123 | No |
| 15 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 32783 | -0.819 | -0.8096 | No |
| 16 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33267 | -0.949 | -0.8147 | Yes |
| 17 | CCNB1 | cyclin B1 [Source:HGNC Symbol;Acc:HGNC:1579] | 33479 | -1.012 | -0.8119 | Yes |
| 18 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33690 | -1.077 | -0.8086 | Yes |
| 19 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 33697 | -1.078 | -0.7997 | Yes |
| 20 | TPX2 | TPX2 microtubule nucleation factor [Source:HGNC Symbol;Acc:HGNC:1249] | 33713 | -1.082 | -0.7911 | Yes |
| 21 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.7821 | Yes |
| 22 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 33838 | -1.126 | -0.7758 | Yes |
| 23 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33877 | -1.138 | -0.7673 | Yes |
| 24 | TRAIP | TRAF interacting protein [Source:HGNC Symbol;Acc:HGNC:30764] | 34108 | -1.212 | -0.7634 | Yes |
| 25 | CDC25C | cell division cycle 25C [Source:HGNC Symbol;Acc:HGNC:1727] | 34490 | -1.349 | -0.7624 | Yes |
| 26 | BRD8 | bromodomain containing 8 [Source:HGNC Symbol;Acc:HGNC:19874] | 34513 | -1.359 | -0.7516 | Yes |
| 27 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34522 | -1.361 | -0.7404 | Yes |
| 28 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34533 | -1.364 | -0.7292 | Yes |
| 29 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 34584 | -1.386 | -0.7189 | Yes |
| 30 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34655 | -1.411 | -0.7090 | Yes |
| 31 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34665 | -1.416 | -0.6974 | Yes |
| 32 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34705 | -1.428 | -0.6864 | Yes |
| 33 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 34826 | -1.475 | -0.6773 | Yes |
| 34 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 34860 | -1.492 | -0.6657 | Yes |
| 35 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34896 | -1.510 | -0.6540 | Yes |
| 36 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35131 | -1.642 | -0.6465 | Yes |
| 37 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 35207 | -1.677 | -0.6345 | Yes |
| 38 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35315 | -1.737 | -0.6228 | Yes |
| 39 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 35454 | -1.811 | -0.6113 | Yes |
| 40 | CHAF1A | chromatin assembly factor 1 subunit A [Source:HGNC Symbol;Acc:HGNC:1910] | 35484 | -1.828 | -0.5968 | Yes |
| 41 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35493 | -1.831 | -0.5817 | Yes |
| 42 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35645 | -1.933 | -0.5695 | Yes |
| 43 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 35659 | -1.943 | -0.5536 | Yes |
| 44 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35748 | -2.016 | -0.5391 | Yes |
| 45 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 35799 | -2.059 | -0.5231 | Yes |
| 46 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35912 | -2.142 | -0.5082 | Yes |
| 47 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35930 | -2.153 | -0.4906 | Yes |
| 48 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 35935 | -2.155 | -0.4727 | Yes |
| 49 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35959 | -2.181 | -0.4550 | Yes |
| 50 | STIL | STIL centriolar assembly protein [Source:HGNC Symbol;Acc:HGNC:10879] | 35983 | -2.196 | -0.4372 | Yes |
| 51 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 35988 | -2.197 | -0.4189 | Yes |
| 52 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36062 | -2.250 | -0.4020 | Yes |
| 53 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36081 | -2.263 | -0.3835 | Yes |
| 54 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 36186 | -2.355 | -0.3666 | Yes |
| 55 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36191 | -2.357 | -0.3469 | Yes |
| 56 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36232 | -2.394 | -0.3279 | Yes |
| 57 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36286 | -2.451 | -0.3088 | Yes |
| 58 | PTP4A3 | protein tyrosine phosphatase 4A3 [Source:HGNC Symbol;Acc:HGNC:9636] | 36321 | -2.496 | -0.2888 | Yes |
| 59 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 36455 | -2.653 | -0.2701 | Yes |
| 60 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36470 | -2.674 | -0.2481 | Yes |
| 61 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36505 | -2.720 | -0.2262 | Yes |
| 62 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36546 | -2.789 | -0.2039 | Yes |
| 63 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36620 | -2.903 | -0.1815 | Yes |
| 64 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36643 | -2.941 | -0.1575 | Yes |
| 65 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36649 | -2.950 | -0.1329 | Yes |
| 66 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36696 | -3.045 | -0.1086 | Yes |
| 67 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36733 | -3.106 | -0.0835 | Yes |
| 68 | OIP5 | Opa interacting protein 5 [Source:HGNC Symbol;Acc:HGNC:20300] | 36858 | -3.461 | -0.0578 | Yes |
| 69 | CHEK2 | checkpoint kinase 2 [Source:HGNC Symbol;Acc:HGNC:16627] | 36870 | -3.488 | -0.0289 | Yes |
| 70 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37029 | -4.153 | 0.0016 | Yes |