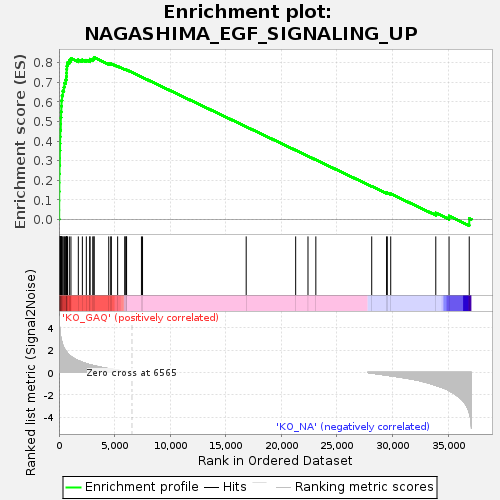
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | NAGASHIMA_EGF_SIGNALING_UP |
| Enrichment Score (ES) | 0.82688993 |
| Normalized Enrichment Score (NES) | 1.90686 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0022193596 |
| FWER p-Value | 0.023 |

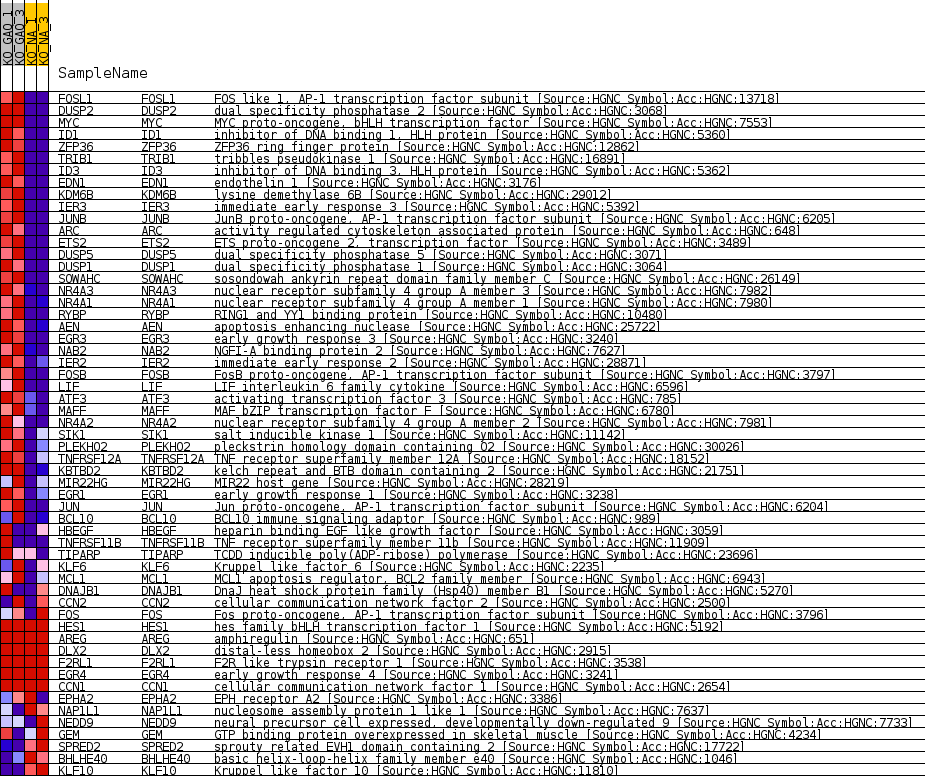
| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FOSL1 | FOS like 1, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:13718] | 15 | 4.678 | 0.0477 | Yes |
| 2 | DUSP2 | dual specificity phosphatase 2 [Source:HGNC Symbol;Acc:HGNC:3068] | 16 | 4.622 | 0.0952 | Yes |
| 3 | MYC | MYC proto-oncogene, bHLH transcription factor [Source:HGNC Symbol;Acc:HGNC:7553] | 18 | 4.578 | 0.1422 | Yes |
| 4 | ID1 | inhibitor of DNA binding 1, HLH protein [Source:HGNC Symbol;Acc:HGNC:5360] | 27 | 4.439 | 0.1876 | Yes |
| 5 | ZFP36 | ZFP36 ring finger protein [Source:HGNC Symbol;Acc:HGNC:12862] | 29 | 4.417 | 0.2330 | Yes |
| 6 | TRIB1 | tribbles pseudokinase 1 [Source:HGNC Symbol;Acc:HGNC:16891] | 48 | 4.124 | 0.2749 | Yes |
| 7 | ID3 | inhibitor of DNA binding 3, HLH protein [Source:HGNC Symbol;Acc:HGNC:5362] | 75 | 3.737 | 0.3126 | Yes |
| 8 | EDN1 | endothelin 1 [Source:HGNC Symbol;Acc:HGNC:3176] | 76 | 3.735 | 0.3510 | Yes |
| 9 | KDM6B | lysine demethylase 6B [Source:HGNC Symbol;Acc:HGNC:29012] | 94 | 3.534 | 0.3869 | Yes |
| 10 | IER3 | immediate early response 3 [Source:HGNC Symbol;Acc:HGNC:5392] | 102 | 3.437 | 0.4220 | Yes |
| 11 | JUNB | JunB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6205] | 116 | 3.363 | 0.4562 | Yes |
| 12 | ARC | activity regulated cytoskeleton associated protein [Source:HGNC Symbol;Acc:HGNC:648] | 159 | 3.121 | 0.4872 | Yes |
| 13 | ETS2 | ETS proto-oncogene 2, transcription factor [Source:HGNC Symbol;Acc:HGNC:3489] | 162 | 3.115 | 0.5191 | Yes |
| 14 | DUSP5 | dual specificity phosphatase 5 [Source:HGNC Symbol;Acc:HGNC:3071] | 188 | 2.968 | 0.5489 | Yes |
| 15 | DUSP1 | dual specificity phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:3064] | 211 | 2.870 | 0.5779 | Yes |
| 16 | SOWAHC | sosondowah ankyrin repeat domain family member C [Source:HGNC Symbol;Acc:HGNC:26149] | 228 | 2.806 | 0.6063 | Yes |
| 17 | NR4A3 | nuclear receptor subfamily 4 group A member 3 [Source:HGNC Symbol;Acc:HGNC:7982] | 273 | 2.659 | 0.6324 | Yes |
| 18 | NR4A1 | nuclear receptor subfamily 4 group A member 1 [Source:HGNC Symbol;Acc:HGNC:7980] | 341 | 2.401 | 0.6553 | Yes |
| 19 | RYBP | RING1 and YY1 binding protein [Source:HGNC Symbol;Acc:HGNC:10480] | 439 | 2.180 | 0.6751 | Yes |
| 20 | AEN | apoptosis enhancing nuclease [Source:HGNC Symbol;Acc:HGNC:25722] | 485 | 2.109 | 0.6955 | Yes |
| 21 | EGR3 | early growth response 3 [Source:HGNC Symbol;Acc:HGNC:3240] | 593 | 1.932 | 0.7125 | Yes |
| 22 | NAB2 | NGFI-A binding protein 2 [Source:HGNC Symbol;Acc:HGNC:7627] | 662 | 1.834 | 0.7295 | Yes |
| 23 | IER2 | immediate early response 2 [Source:HGNC Symbol;Acc:HGNC:28871] | 677 | 1.824 | 0.7479 | Yes |
| 24 | FOSB | FosB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3797] | 688 | 1.811 | 0.7662 | Yes |
| 25 | LIF | LIF interleukin 6 family cytokine [Source:HGNC Symbol;Acc:HGNC:6596] | 711 | 1.779 | 0.7839 | Yes |
| 26 | ATF3 | activating transcription factor 3 [Source:HGNC Symbol;Acc:HGNC:785] | 764 | 1.712 | 0.8001 | Yes |
| 27 | MAFF | MAF bZIP transcription factor F [Source:HGNC Symbol;Acc:HGNC:6780] | 928 | 1.535 | 0.8115 | Yes |
| 28 | NR4A2 | nuclear receptor subfamily 4 group A member 2 [Source:HGNC Symbol;Acc:HGNC:7981] | 1074 | 1.414 | 0.8221 | Yes |
| 29 | SIK1 | salt inducible kinase 1 [Source:HGNC Symbol;Acc:HGNC:11142] | 1738 | 1.044 | 0.8149 | Yes |
| 30 | PLEKHO2 | pleckstrin homology domain containing O2 [Source:HGNC Symbol;Acc:HGNC:30026] | 2102 | 0.894 | 0.8143 | Yes |
| 31 | TNFRSF12A | TNF receptor superfamily member 12A [Source:HGNC Symbol;Acc:HGNC:18152] | 2458 | 0.778 | 0.8127 | Yes |
| 32 | KBTBD2 | kelch repeat and BTB domain containing 2 [Source:HGNC Symbol;Acc:HGNC:21751] | 2768 | 0.690 | 0.8115 | Yes |
| 33 | MIR22HG | MIR22 host gene [Source:HGNC Symbol;Acc:HGNC:28219] | 2787 | 0.685 | 0.8180 | Yes |
| 34 | EGR1 | early growth response 1 [Source:HGNC Symbol;Acc:HGNC:3238] | 3036 | 0.622 | 0.8177 | Yes |
| 35 | JUN | Jun proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6204] | 3144 | 0.596 | 0.8210 | Yes |
| 36 | BCL10 | BCL10 immune signaling adaptor [Source:HGNC Symbol;Acc:HGNC:989] | 3152 | 0.595 | 0.8269 | Yes |
| 37 | HBEGF | heparin binding EGF like growth factor [Source:HGNC Symbol;Acc:HGNC:3059] | 4474 | 0.332 | 0.7946 | No |
| 38 | TNFRSF11B | TNF receptor superfamily member 11b [Source:HGNC Symbol;Acc:HGNC:11909] | 4652 | 0.304 | 0.7930 | No |
| 39 | TIPARP | TCDD inducible poly(ADP-ribose) polymerase [Source:HGNC Symbol;Acc:HGNC:23696] | 4706 | 0.295 | 0.7946 | No |
| 40 | KLF6 | Kruppel like factor 6 [Source:HGNC Symbol;Acc:HGNC:2235] | 5274 | 0.196 | 0.7813 | No |
| 41 | MCL1 | MCL1 apoptosis regulator, BCL2 family member [Source:HGNC Symbol;Acc:HGNC:6943] | 5924 | 0.102 | 0.7648 | No |
| 42 | DNAJB1 | DnaJ heat shock protein family (Hsp40) member B1 [Source:HGNC Symbol;Acc:HGNC:5270] | 5942 | 0.099 | 0.7654 | No |
| 43 | CCN2 | cellular communication network factor 2 [Source:HGNC Symbol;Acc:HGNC:2500] | 6040 | 0.087 | 0.7636 | No |
| 44 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796] | 6078 | 0.083 | 0.7635 | No |
| 45 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] | 7415 | 0.000 | 0.7274 | No |
| 46 | AREG | amphiregulin [Source:HGNC Symbol;Acc:HGNC:651] | 7513 | 0.000 | 0.7248 | No |
| 47 | DLX2 | distal-less homeobox 2 [Source:HGNC Symbol;Acc:HGNC:2915] | 16836 | 0.000 | 0.4731 | No |
| 48 | F2RL1 | F2R like trypsin receptor 1 [Source:HGNC Symbol;Acc:HGNC:3538] | 21286 | 0.000 | 0.3530 | No |
| 49 | EGR4 | early growth response 4 [Source:HGNC Symbol;Acc:HGNC:3241] | 22394 | 0.000 | 0.3231 | No |
| 50 | CCN1 | cellular communication network factor 1 [Source:HGNC Symbol;Acc:HGNC:2654] | 23100 | 0.000 | 0.3040 | No |
| 51 | EPHA2 | EPH receptor A2 [Source:HGNC Symbol;Acc:HGNC:3386] | 28126 | -0.052 | 0.1689 | No |
| 52 | NAP1L1 | nucleosome assembly protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:7637] | 29462 | -0.239 | 0.1353 | No |
| 53 | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 [Source:HGNC Symbol;Acc:HGNC:7733] | 29527 | -0.245 | 0.1361 | No |
| 54 | GEM | GTP binding protein overexpressed in skeletal muscle [Source:HGNC Symbol;Acc:HGNC:4234] | 29835 | -0.294 | 0.1308 | No |
| 55 | SPRED2 | sprouty related EVH1 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:17722] | 33888 | -1.142 | 0.0331 | No |
| 56 | BHLHE40 | basic helix-loop-helix family member e40 [Source:HGNC Symbol;Acc:HGNC:1046] | 35081 | -1.617 | 0.0176 | No |
| 57 | KLF10 | Kruppel like factor 10 [Source:HGNC Symbol;Acc:HGNC:11810] | 36897 | -3.566 | 0.0052 | No |