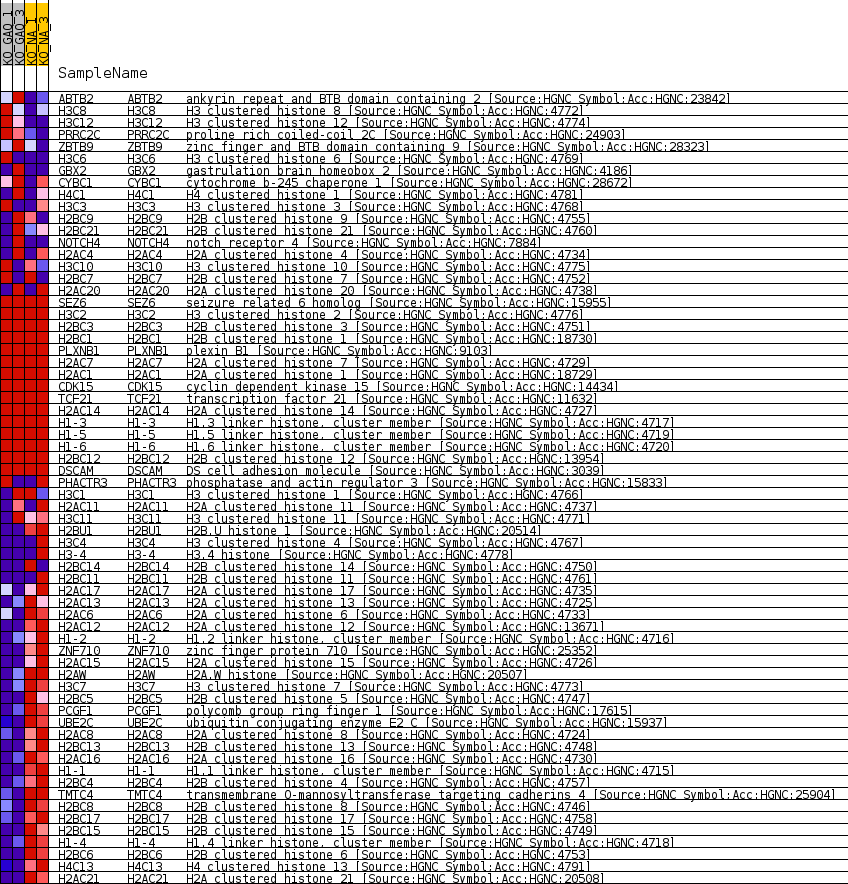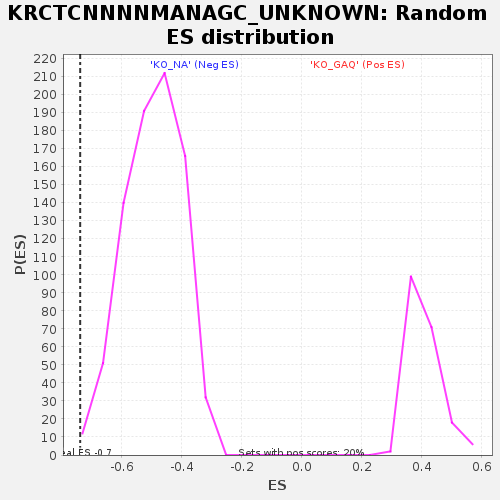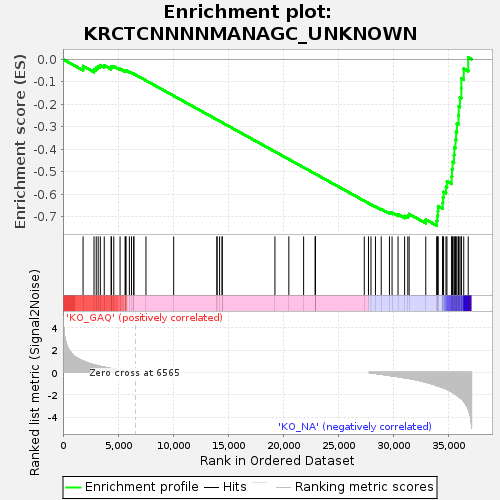
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | KRCTCNNNNMANAGC_UNKNOWN |
| Enrichment Score (ES) | -0.7391287 |
| Normalized Enrichment Score (NES) | -1.4892136 |
| Nominal p-value | 0.0024875621 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.992 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ABTB2 | ankyrin repeat and BTB domain containing 2 [Source:HGNC Symbol;Acc:HGNC:23842] | 1821 | 1.007 | -0.0301 | No |
| 2 | H3C8 | H3 clustered histone 8 [Source:HGNC Symbol;Acc:HGNC:4772] | 2815 | 0.679 | -0.0440 | No |
| 3 | H3C12 | H3 clustered histone 12 [Source:HGNC Symbol;Acc:HGNC:4774] | 3016 | 0.626 | -0.0376 | No |
| 4 | PRRC2C | proline rich coiled-coil 2C [Source:HGNC Symbol;Acc:HGNC:24903] | 3193 | 0.585 | -0.0312 | No |
| 5 | ZBTB9 | zinc finger and BTB domain containing 9 [Source:HGNC Symbol;Acc:HGNC:28323] | 3388 | 0.540 | -0.0263 | No |
| 6 | H3C6 | H3 clustered histone 6 [Source:HGNC Symbol;Acc:HGNC:4769] | 3744 | 0.463 | -0.0271 | No |
| 7 | GBX2 | gastrulation brain homeobox 2 [Source:HGNC Symbol;Acc:HGNC:4186] | 4367 | 0.349 | -0.0373 | No |
| 8 | CYBC1 | cytochrome b-245 chaperone 1 [Source:HGNC Symbol;Acc:HGNC:28672] | 4398 | 0.344 | -0.0316 | No |
| 9 | H4C1 | H4 clustered histone 1 [Source:HGNC Symbol;Acc:HGNC:4781] | 4610 | 0.310 | -0.0314 | No |
| 10 | H3C3 | H3 clustered histone 3 [Source:HGNC Symbol;Acc:HGNC:4768] | 5180 | 0.210 | -0.0428 | No |
| 11 | H2BC9 | H2B clustered histone 9 [Source:HGNC Symbol;Acc:HGNC:4755] | 5636 | 0.140 | -0.0524 | No |
| 12 | H2BC21 | H2B clustered histone 21 [Source:HGNC Symbol;Acc:HGNC:4760] | 5679 | 0.140 | -0.0509 | No |
| 13 | NOTCH4 | notch receptor 4 [Source:HGNC Symbol;Acc:HGNC:7884] | 5722 | 0.134 | -0.0495 | No |
| 14 | H2AC4 | H2A clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4734] | 6025 | 0.087 | -0.0560 | No |
| 15 | H3C10 | H3 clustered histone 10 [Source:HGNC Symbol;Acc:HGNC:4775] | 6221 | 0.057 | -0.0602 | No |
| 16 | H2BC7 | H2B clustered histone 7 [Source:HGNC Symbol;Acc:HGNC:4752] | 6423 | 0.026 | -0.0651 | No |
| 17 | H2AC20 | H2A clustered histone 20 [Source:HGNC Symbol;Acc:HGNC:4738] | 6432 | 0.025 | -0.0649 | No |
| 18 | SEZ6 | seizure related 6 homolog [Source:HGNC Symbol;Acc:HGNC:15955] | 7531 | 0.000 | -0.0945 | No |
| 19 | H3C2 | H3 clustered histone 2 [Source:HGNC Symbol;Acc:HGNC:4776] | 10047 | 0.000 | -0.1624 | No |
| 20 | H2BC3 | H2B clustered histone 3 [Source:HGNC Symbol;Acc:HGNC:4751] | 13977 | 0.000 | -0.2686 | No |
| 21 | H2BC1 | H2B clustered histone 1 [Source:HGNC Symbol;Acc:HGNC:18730] | 13980 | 0.000 | -0.2686 | No |
| 22 | PLXNB1 | plexin B1 [Source:HGNC Symbol;Acc:HGNC:9103] | 14215 | 0.000 | -0.2749 | No |
| 23 | H2AC7 | H2A clustered histone 7 [Source:HGNC Symbol;Acc:HGNC:4729] | 14447 | 0.000 | -0.2812 | No |
| 24 | H2AC1 | H2A clustered histone 1 [Source:HGNC Symbol;Acc:HGNC:18729] | 14450 | 0.000 | -0.2812 | No |
| 25 | CDK15 | cyclin dependent kinase 15 [Source:HGNC Symbol;Acc:HGNC:14434] | 19238 | 0.000 | -0.4105 | No |
| 26 | TCF21 | transcription factor 21 [Source:HGNC Symbol;Acc:HGNC:11632] | 20501 | 0.000 | -0.4446 | No |
| 27 | H2AC14 | H2A clustered histone 14 [Source:HGNC Symbol;Acc:HGNC:4727] | 21843 | 0.000 | -0.4808 | No |
| 28 | H1-3 | H1.3 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4717] | 22905 | 0.000 | -0.5095 | No |
| 29 | H1-5 | H1.5 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4719] | 22906 | 0.000 | -0.5095 | No |
| 30 | H1-6 | H1.6 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4720] | 22910 | 0.000 | -0.5095 | No |
| 31 | H2BC12 | H2B clustered histone 12 [Source:HGNC Symbol;Acc:HGNC:13954] | 27349 | 0.000 | -0.6294 | No |
| 32 | DSCAM | DS cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:3039] | 27731 | 0.000 | -0.6397 | No |
| 33 | PHACTR3 | phosphatase and actin regulator 3 [Source:HGNC Symbol;Acc:HGNC:15833] | 27960 | -0.029 | -0.6453 | No |
| 34 | H3C1 | H3 clustered histone 1 [Source:HGNC Symbol;Acc:HGNC:4766] | 28365 | -0.085 | -0.6546 | No |
| 35 | H2AC11 | H2A clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4737] | 28889 | -0.155 | -0.6658 | No |
| 36 | H3C11 | H3 clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4771] | 29636 | -0.263 | -0.6810 | No |
| 37 | H2BU1 | H2B.U histone 1 [Source:HGNC Symbol;Acc:HGNC:20514] | 29872 | -0.300 | -0.6816 | No |
| 38 | H3C4 | H3 clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4767] | 30415 | -0.371 | -0.6893 | No |
| 39 | H3-4 | H3.4 histone [Source:HGNC Symbol;Acc:HGNC:4778] | 31013 | -0.464 | -0.6966 | No |
| 40 | H2BC14 | H2B clustered histone 14 [Source:HGNC Symbol;Acc:HGNC:4750] | 31302 | -0.510 | -0.6947 | No |
| 41 | H2BC11 | H2B clustered histone 11 [Source:HGNC Symbol;Acc:HGNC:4761] | 31423 | -0.531 | -0.6879 | No |
| 42 | H2AC17 | H2A clustered histone 17 [Source:HGNC Symbol;Acc:HGNC:4735] | 32932 | -0.855 | -0.7124 | No |
| 43 | H2AC13 | H2A clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:4725] | 33922 | -1.158 | -0.7172 | Yes |
| 44 | H2AC6 | H2A clustered histone 6 [Source:HGNC Symbol;Acc:HGNC:4733] | 33978 | -1.178 | -0.6964 | Yes |
| 45 | H2AC12 | H2A clustered histone 12 [Source:HGNC Symbol;Acc:HGNC:13671] | 34033 | -1.195 | -0.6752 | Yes |
| 46 | H1-2 | H1.2 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4716] | 34048 | -1.199 | -0.6528 | Yes |
| 47 | ZNF710 | zinc finger protein 710 [Source:HGNC Symbol;Acc:HGNC:25352] | 34456 | -1.337 | -0.6385 | Yes |
| 48 | H2AC15 | H2A clustered histone 15 [Source:HGNC Symbol;Acc:HGNC:4726] | 34492 | -1.349 | -0.6139 | Yes |
| 49 | H2AW | H2A.W histone [Source:HGNC Symbol;Acc:HGNC:20507] | 34548 | -1.367 | -0.5895 | Yes |
| 50 | H3C7 | H3 clustered histone 7 [Source:HGNC Symbol;Acc:HGNC:4773] | 34767 | -1.449 | -0.5679 | Yes |
| 51 | H2BC5 | H2B clustered histone 5 [Source:HGNC Symbol;Acc:HGNC:4747] | 34853 | -1.486 | -0.5421 | Yes |
| 52 | PCGF1 | polycomb group ring finger 1 [Source:HGNC Symbol;Acc:HGNC:17615] | 35278 | -1.719 | -0.5209 | Yes |
| 53 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35315 | -1.737 | -0.4890 | Yes |
| 54 | H2AC8 | H2A clustered histone 8 [Source:HGNC Symbol;Acc:HGNC:4724] | 35363 | -1.759 | -0.4569 | Yes |
| 55 | H2BC13 | H2B clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:4748] | 35492 | -1.831 | -0.4257 | Yes |
| 56 | H2AC16 | H2A clustered histone 16 [Source:HGNC Symbol;Acc:HGNC:4730] | 35523 | -1.853 | -0.3914 | Yes |
| 57 | H1-1 | H1.1 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4715] | 35628 | -1.922 | -0.3578 | Yes |
| 58 | H2BC4 | H2B clustered histone 4 [Source:HGNC Symbol;Acc:HGNC:4757] | 35683 | -1.962 | -0.3221 | Yes |
| 59 | TMTC4 | transmembrane O-mannosyltransferase targeting cadherins 4 [Source:HGNC Symbol;Acc:HGNC:25904] | 35762 | -2.027 | -0.2858 | Yes |
| 60 | H2BC8 | H2B clustered histone 8 [Source:HGNC Symbol;Acc:HGNC:4746] | 35899 | -2.127 | -0.2492 | Yes |
| 61 | H2BC17 | H2B clustered histone 17 [Source:HGNC Symbol;Acc:HGNC:4758] | 35919 | -2.148 | -0.2090 | Yes |
| 62 | H2BC15 | H2B clustered histone 15 [Source:HGNC Symbol;Acc:HGNC:4749] | 36032 | -2.228 | -0.1699 | Yes |
| 63 | H1-4 | H1.4 linker histone, cluster member [Source:HGNC Symbol;Acc:HGNC:4718] | 36160 | -2.338 | -0.1290 | Yes |
| 64 | H2BC6 | H2B clustered histone 6 [Source:HGNC Symbol;Acc:HGNC:4753] | 36164 | -2.341 | -0.0847 | Yes |
| 65 | H4C13 | H4 clustered histone 13 [Source:HGNC Symbol;Acc:HGNC:4791] | 36379 | -2.561 | -0.0420 | Yes |
| 66 | H2AC21 | H2A clustered histone 21 [Source:HGNC Symbol;Acc:HGNC:20508] | 36785 | -3.229 | 0.0082 | Yes |