Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
| GeneSet | MIR1911_5P |
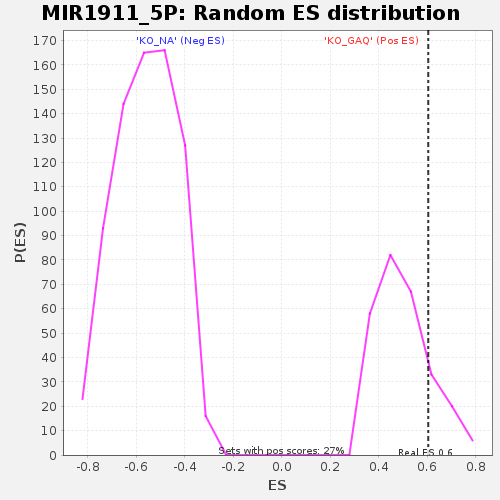
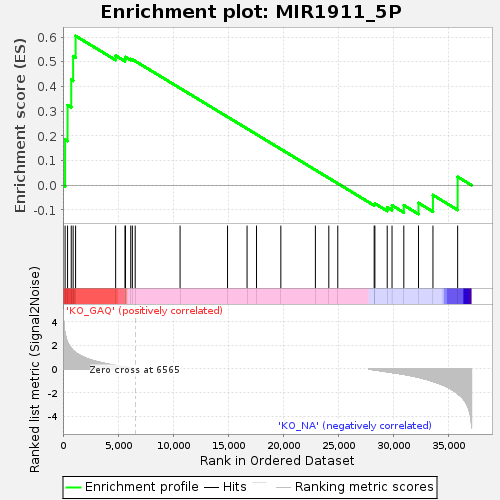
| Enrichment Score (ES) | 0.6049871 |
| Normalized Enrichment Score (NES) | 1.2098956 |
| Nominal p-value | 0.19172932 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EIF2AK2 | eukaryotic translation initiation factor 2 alpha kinase 2 [Source:HGNC Symbol;Acc:HGNC:9437] | 191 | 2.942 | 0.1852 | Yes |
| 2 | SUSD6 | sushi domain containing 6 [Source:HGNC Symbol;Acc:HGNC:19956] | 414 | 2.242 | 0.3243 | Yes |
| 3 | GJA1 | gap junction protein alpha 1 [Source:HGNC Symbol;Acc:HGNC:4274] | 748 | 1.732 | 0.4274 | Yes |
| 4 | RASA2 | RAS p21 protein activator 2 [Source:HGNC Symbol;Acc:HGNC:9872] | 920 | 1.541 | 0.5226 | Yes |
| 5 | LMTK2 | lemur tyrosine kinase 2 [Source:HGNC Symbol;Acc:HGNC:17880] | 1143 | 1.366 | 0.6050 | Yes |
| 6 | B3GNT2 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:15629] | 4779 | 0.282 | 0.5252 | No |
| 7 | G3BP2 | G3BP stress granule assembly factor 2 [Source:HGNC Symbol;Acc:HGNC:30291] | 5628 | 0.142 | 0.5115 | No |
| 8 | ZNF93 | zinc finger protein 93 [Source:HGNC Symbol;Acc:HGNC:13169] | 5672 | 0.140 | 0.5194 | No |
| 9 | NKAPD1 | NKAP domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25569] | 6150 | 0.070 | 0.5111 | No |
| 10 | CSMD3 | CUB and Sushi multiple domains 3 [Source:HGNC Symbol;Acc:HGNC:19291] | 6296 | 0.042 | 0.5099 | No |
| 11 | SERF2 | small EDRK-rich factor 2 [Source:HGNC Symbol;Acc:HGNC:10757] | 6554 | 0.001 | 0.5030 | No |
| 12 | CRIP3 | cysteine rich protein 3 [Source:HGNC Symbol;Acc:HGNC:17751] | 10628 | 0.000 | 0.3932 | No |
| 13 | SYTL4 | synaptotagmin like 4 [Source:HGNC Symbol;Acc:HGNC:15588] | 14937 | 0.000 | 0.2769 | No |
| 14 | DLC1 | DLC1 Rho GTPase activating protein [Source:HGNC Symbol;Acc:HGNC:2897] | 16709 | 0.000 | 0.2291 | No |
| 15 | GPR45 | G protein-coupled receptor 45 [Source:HGNC Symbol;Acc:HGNC:4503] | 17568 | 0.000 | 0.2060 | No |
| 16 | SMARCA1 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 [Source:HGNC Symbol;Acc:HGNC:11097] | 19775 | 0.000 | 0.1465 | No |
| 17 | FANCD2OS | FANCD2 opposite strand [Source:HGNC Symbol;Acc:HGNC:28623] | 22911 | 0.000 | 0.0619 | No |
| 18 | DISC1 | DISC1 scaffold protein [Source:HGNC Symbol;Acc:HGNC:2888] | 24130 | 0.000 | 0.0290 | No |
| 19 | KRTAP24-1 | keratin associated protein 24-1 [Source:HGNC Symbol;Acc:HGNC:33902] | 24944 | 0.000 | 0.0071 | No |
| 20 | ARHGEF33 | Rho guanine nucleotide exchange factor 33 [Source:HGNC Symbol;Acc:HGNC:37252] | 28248 | -0.070 | -0.0775 | No |
| 21 | FUT8 | fucosyltransferase 8 [Source:HGNC Symbol;Acc:HGNC:4019] | 28313 | -0.079 | -0.0741 | No |
| 22 | C6orf62 | chromosome 6 open reading frame 62 [Source:HGNC Symbol;Acc:HGNC:20998] | 29432 | -0.236 | -0.0891 | No |
| 23 | FRS2 | fibroblast growth factor receptor substrate 2 [Source:HGNC Symbol;Acc:HGNC:16971] | 29869 | -0.299 | -0.0815 | No |
| 24 | BICD2 | BICD cargo adaptor 2 [Source:HGNC Symbol;Acc:HGNC:17208] | 30938 | -0.454 | -0.0809 | No |
| 25 | DERA | deoxyribose-phosphate aldolase [Source:HGNC Symbol;Acc:HGNC:24269] | 32271 | -0.697 | -0.0717 | No |
| 26 | PDIK1L | PDLIM1 interacting kinase 1 like [Source:HGNC Symbol;Acc:HGNC:18981] | 33584 | -1.043 | -0.0396 | No |
| 27 | CTC1 | CST telomere replication complex component 1 [Source:HGNC Symbol;Acc:HGNC:26169] | 35826 | -2.073 | 0.0341 | No |