Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR5088_5P |
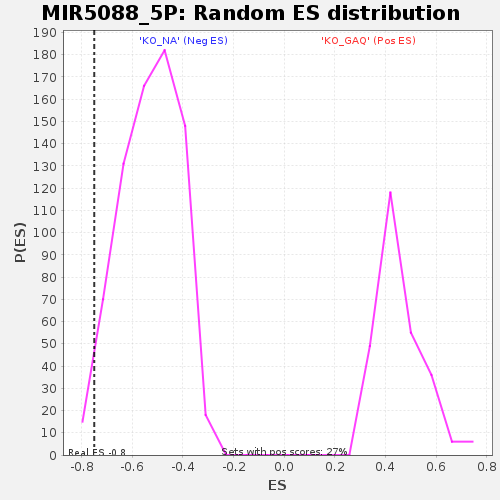
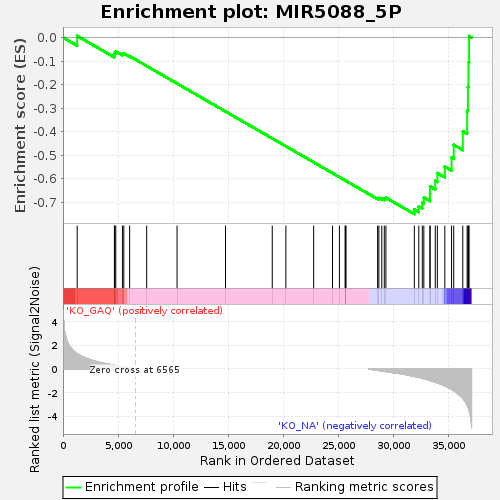
| Enrichment Score (ES) | -0.75022835 |
| Normalized Enrichment Score (NES) | -1.4203827 |
| Nominal p-value | 0.026027398 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | USP14 | ubiquitin specific peptidase 14 [Source:HGNC Symbol;Acc:HGNC:12612] | 1289 | 1.280 | 0.0066 | No |
| 2 | HIC2 | HIC ZBTB transcriptional repressor 2 [Source:HGNC Symbol;Acc:HGNC:18595] | 4666 | 0.302 | -0.0747 | No |
| 3 | MSANTD2 | Myb/SANT DNA binding domain containing 2 [Source:HGNC Symbol;Acc:HGNC:26266] | 4695 | 0.298 | -0.0658 | No |
| 4 | ZNF135 | zinc finger protein 135 [Source:HGNC Symbol;Acc:HGNC:12919] | 4789 | 0.280 | -0.0593 | No |
| 5 | HECW2 | HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:29853] | 5396 | 0.177 | -0.0699 | No |
| 6 | KDM2A | lysine demethylase 2A [Source:HGNC Symbol;Acc:HGNC:13606] | 5520 | 0.158 | -0.0681 | No |
| 7 | CTDNEP1 | CTD nuclear envelope phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:19085] | 6048 | 0.087 | -0.0795 | No |
| 8 | CNTNAP2 | contactin associated protein 2 [Source:HGNC Symbol;Acc:HGNC:13830] | 7596 | 0.000 | -0.1213 | No |
| 9 | SLC22A2 | solute carrier family 22 member 2 [Source:HGNC Symbol;Acc:HGNC:10966] | 10354 | 0.000 | -0.1957 | No |
| 10 | PCDH10 | protocadherin 10 [Source:HGNC Symbol;Acc:HGNC:13404] | 14752 | 0.000 | -0.3143 | No |
| 11 | MIP | major intrinsic protein of lens fiber [Source:HGNC Symbol;Acc:HGNC:7103] | 18996 | 0.000 | -0.4289 | No |
| 12 | TMEM47 | transmembrane protein 47 [Source:HGNC Symbol;Acc:HGNC:18515] | 20232 | 0.000 | -0.4622 | No |
| 13 | PI15 | peptidase inhibitor 15 [Source:HGNC Symbol;Acc:HGNC:8946] | 22755 | 0.000 | -0.5302 | No |
| 14 | ADCYAP1R1 | ADCYAP receptor type I [Source:HGNC Symbol;Acc:HGNC:242] | 24466 | 0.000 | -0.5764 | No |
| 15 | KCNMB2 | potassium calcium-activated channel subfamily M regulatory beta subunit 2 [Source:HGNC Symbol;Acc:HGNC:6286] | 25092 | 0.000 | -0.5933 | No |
| 16 | PRSS8 | serine protease 8 [Source:HGNC Symbol;Acc:HGNC:9491] | 25608 | 0.000 | -0.6072 | No |
| 17 | RIMS4 | regulating synaptic membrane exocytosis 4 [Source:HGNC Symbol;Acc:HGNC:16183] | 25672 | 0.000 | -0.6089 | No |
| 18 | FGF9 | fibroblast growth factor 9 [Source:HGNC Symbol;Acc:HGNC:3687] | 25676 | 0.000 | -0.6089 | No |
| 19 | AKAP3 | A-kinase anchoring protein 3 [Source:HGNC Symbol;Acc:HGNC:373] | 28560 | -0.112 | -0.6832 | No |
| 20 | MECP2 | methyl-CpG binding protein 2 [Source:HGNC Symbol;Acc:HGNC:6990] | 28673 | -0.129 | -0.6820 | No |
| 21 | LRRTM2 | leucine rich repeat transmembrane neuronal 2 [Source:HGNC Symbol;Acc:HGNC:19409] | 28938 | -0.163 | -0.6839 | No |
| 22 | CANX | calnexin [Source:HGNC Symbol;Acc:HGNC:1473] | 29167 | -0.195 | -0.6837 | No |
| 23 | PSMB9 | proteasome 20S subunit beta 9 [Source:HGNC Symbol;Acc:HGNC:9546] | 29314 | -0.217 | -0.6806 | No |
| 24 | BZW2 | basic leucine zipper and W2 domains 2 [Source:HGNC Symbol;Acc:HGNC:18808] | 31894 | -0.615 | -0.7303 | Yes |
| 25 | PJA1 | praja ring finger ubiquitin ligase 1 [Source:HGNC Symbol;Acc:HGNC:16648] | 32299 | -0.703 | -0.7185 | Yes |
| 26 | ATPAF1 | ATP synthase mitochondrial F1 complex assembly factor 1 [Source:HGNC Symbol;Acc:HGNC:18803] | 32617 | -0.775 | -0.7020 | Yes |
| 27 | VAPB | VAMP associated protein B and C [Source:HGNC Symbol;Acc:HGNC:12649] | 32756 | -0.814 | -0.6793 | Yes |
| 28 | XRCC6 | X-ray repair cross complementing 6 [Source:HGNC Symbol;Acc:HGNC:4055] | 33316 | -0.961 | -0.6633 | Yes |
| 29 | PTER | phosphotriesterase related [Source:HGNC Symbol;Acc:HGNC:9590] | 33322 | -0.963 | -0.6323 | Yes |
| 30 | PTBP2 | polypyrimidine tract binding protein 2 [Source:HGNC Symbol;Acc:HGNC:17662] | 33794 | -1.111 | -0.6090 | Yes |
| 31 | GALNT2 | polypeptide N-acetylgalactosaminyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:4124] | 33982 | -1.180 | -0.5759 | Yes |
| 32 | THTPA | thiamine triphosphatase [Source:HGNC Symbol;Acc:HGNC:18987] | 34662 | -1.414 | -0.5485 | Yes |
| 33 | CENPI | centromere protein I [Source:HGNC Symbol;Acc:HGNC:3968] | 35272 | -1.715 | -0.5094 | Yes |
| 34 | IDS | iduronate 2-sulfatase [Source:HGNC Symbol;Acc:HGNC:5389] | 35468 | -1.816 | -0.4559 | Yes |
| 35 | FOXO4 | forkhead box O4 [Source:HGNC Symbol;Acc:HGNC:7139] | 36293 | -2.461 | -0.3984 | Yes |
| 36 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 [Source:HGNC Symbol;Acc:HGNC:16922] | 36688 | -3.028 | -0.3111 | Yes |
| 37 | PMP22 | peripheral myelin protein 22 [Source:HGNC Symbol;Acc:HGNC:9118] | 36760 | -3.182 | -0.2100 | Yes |
| 38 | FUT10 | fucosyltransferase 10 [Source:HGNC Symbol;Acc:HGNC:19234] | 36807 | -3.288 | -0.1049 | Yes |
| 39 | SH3PXD2A | SH3 and PX domains 2A [Source:HGNC Symbol;Acc:HGNC:23664] | 36865 | -3.476 | 0.0061 | Yes |