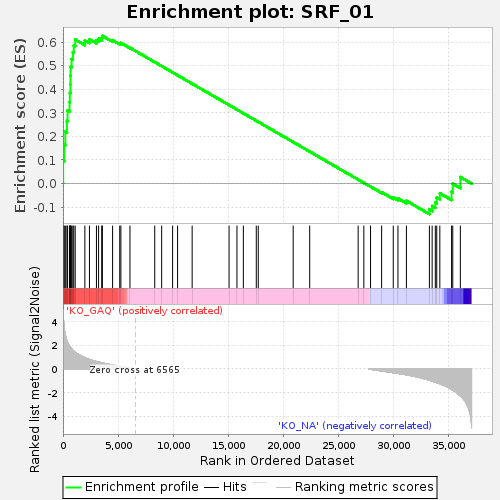
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_GAQ |
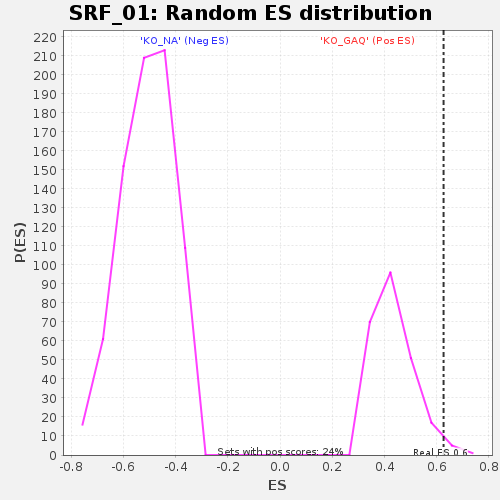
| GeneSet | SRF_01 |
| Enrichment Score (ES) | 0.6262349 |
| Normalized Enrichment Score (NES) | 1.4383515 |
| Nominal p-value | 0.025 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FOSL1 | FOS like 1, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:13718] | 15 | 4.678 | 0.0965 | Yes |
| 2 | JUNB | JunB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:6205] | 116 | 3.363 | 0.1635 | Yes |
| 3 | ACTG2 | actin gamma 2, smooth muscle [Source:HGNC Symbol;Acc:HGNC:145] | 212 | 2.863 | 0.2202 | Yes |
| 4 | ACTN1 | actinin alpha 1 [Source:HGNC Symbol;Acc:HGNC:163] | 354 | 2.372 | 0.2655 | Yes |
| 5 | WDR1 | WD repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:12754] | 416 | 2.241 | 0.3103 | Yes |
| 6 | EGR3 | early growth response 3 [Source:HGNC Symbol;Acc:HGNC:3240] | 593 | 1.932 | 0.3456 | Yes |
| 7 | HOXB4 | homeobox B4 [Source:HGNC Symbol;Acc:HGNC:5115] | 621 | 1.890 | 0.3840 | Yes |
| 8 | IER2 | immediate early response 2 [Source:HGNC Symbol;Acc:HGNC:28871] | 677 | 1.824 | 0.4203 | Yes |
| 9 | FLNA | filamin A [Source:HGNC Symbol;Acc:HGNC:3754] | 681 | 1.820 | 0.4579 | Yes |
| 10 | FOSB | FosB proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3797] | 688 | 1.811 | 0.4953 | Yes |
| 11 | PFN1 | profilin 1 [Source:HGNC Symbol;Acc:HGNC:8881] | 781 | 1.686 | 0.5277 | Yes |
| 12 | CFL1 | cofilin 1 [Source:HGNC Symbol;Acc:HGNC:1874] | 891 | 1.564 | 0.5572 | Yes |
| 13 | ACTR3 | actin related protein 3 [Source:HGNC Symbol;Acc:HGNC:170] | 978 | 1.490 | 0.5857 | Yes |
| 14 | MYL6 | myosin light chain 6 [Source:HGNC Symbol;Acc:HGNC:7587] | 1107 | 1.391 | 0.6111 | Yes |
| 15 | MYH11 | myosin heavy chain 11 [Source:HGNC Symbol;Acc:HGNC:7569] | 1985 | 0.945 | 0.6070 | Yes |
| 16 | CNN2 | calponin 2 [Source:HGNC Symbol;Acc:HGNC:2156] | 2401 | 0.791 | 0.6121 | Yes |
| 17 | EGR1 | early growth response 1 [Source:HGNC Symbol;Acc:HGNC:3238] | 3036 | 0.622 | 0.6079 | Yes |
| 18 | CAP1 | cyclase associated actin cytoskeleton regulatory protein 1 [Source:HGNC Symbol;Acc:HGNC:20040] | 3231 | 0.576 | 0.6146 | Yes |
| 19 | THBS1 | thrombospondin 1 [Source:HGNC Symbol;Acc:HGNC:11785] | 3513 | 0.510 | 0.6176 | Yes |
| 20 | ASB2 | ankyrin repeat and SOCS box containing 2 [Source:HGNC Symbol;Acc:HGNC:16012] | 3575 | 0.497 | 0.6262 | Yes |
| 21 | ITGB1BP2 | integrin subunit beta 1 binding protein 2 [Source:HGNC Symbol;Acc:HGNC:6154] | 4502 | 0.326 | 0.6080 | No |
| 22 | TNNC1 | troponin C1, slow skeletal and cardiac type [Source:HGNC Symbol;Acc:HGNC:11943] | 5151 | 0.214 | 0.5949 | No |
| 23 | SCOC | short coiled-coil protein [Source:HGNC Symbol;Acc:HGNC:20335] | 5254 | 0.200 | 0.5963 | No |
| 24 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796] | 6078 | 0.083 | 0.5758 | No |
| 25 | CD248 | CD248 molecule [Source:HGNC Symbol;Acc:HGNC:18219] | 8327 | 0.000 | 0.5151 | No |
| 26 | CALD1 | caldesmon 1 [Source:HGNC Symbol;Acc:HGNC:1441] | 8957 | 0.000 | 0.4981 | No |
| 27 | NPAS4 | neuronal PAS domain protein 4 [Source:HGNC Symbol;Acc:HGNC:18983] | 9950 | 0.000 | 0.4714 | No |
| 28 | SUSD1 | sushi domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25413] | 10402 | 0.000 | 0.4592 | No |
| 29 | PRM1 | protamine 1 [Source:HGNC Symbol;Acc:HGNC:9447] | 11724 | 0.000 | 0.4235 | No |
| 30 | TNMD | tenomodulin [Source:HGNC Symbol;Acc:HGNC:17757] | 15074 | 0.000 | 0.3331 | No |
| 31 | CA3 | carbonic anhydrase 3 [Source:HGNC Symbol;Acc:HGNC:1374] | 15792 | 0.000 | 0.3138 | No |
| 32 | PHOX2B | paired like homeobox 2B [Source:HGNC Symbol;Acc:HGNC:9143] | 16373 | 0.000 | 0.2981 | No |
| 33 | GPR20 | G protein-coupled receptor 20 [Source:HGNC Symbol;Acc:HGNC:4475] | 17545 | 0.000 | 0.2665 | No |
| 34 | STX10 | syntaxin 10 [Source:HGNC Symbol;Acc:HGNC:11428] | 17725 | 0.000 | 0.2616 | No |
| 35 | IGF2-AS | IGF2 antisense RNA [Source:HGNC Symbol;Acc:HGNC:14062] | 20896 | 0.000 | 0.1761 | No |
| 36 | EGR4 | early growth response 4 [Source:HGNC Symbol;Acc:HGNC:3241] | 22394 | 0.000 | 0.1356 | No |
| 37 | IRAG1 | inositol 1,4,5-triphosphate receptor associated 1 [Source:HGNC Symbol;Acc:HGNC:7237] | 26793 | 0.000 | 0.0169 | No |
| 38 | IL17B | interleukin 17B [Source:HGNC Symbol;Acc:HGNC:5982] | 27310 | 0.000 | 0.0030 | No |
| 39 | TPM1 | tropomyosin 1 [Source:HGNC Symbol;Acc:HGNC:12010] | 27917 | -0.022 | -0.0129 | No |
| 40 | KCNMB1 | potassium calcium-activated channel subfamily M regulatory beta subunit 1 [Source:HGNC Symbol;Acc:HGNC:6285] | 28925 | -0.161 | -0.0368 | No |
| 41 | PRUNE2 | prune homolog 2 with BCH domain [Source:HGNC Symbol;Acc:HGNC:25209] | 29981 | -0.313 | -0.0588 | No |
| 42 | MUS81 | MUS81 structure-specific endonuclease subunit [Source:HGNC Symbol;Acc:HGNC:29814] | 30407 | -0.370 | -0.0626 | No |
| 43 | TAZ | tafazzin [Source:HGNC Symbol;Acc:HGNC:11577] | 31175 | -0.490 | -0.0731 | No |
| 44 | TGFB1I1 | transforming growth factor beta 1 induced transcript 1 [Source:HGNC Symbol;Acc:HGNC:11767] | 33261 | -0.948 | -0.1098 | No |
| 45 | SETD2 | SET domain containing 2, histone lysine methyltransferase [Source:HGNC Symbol;Acc:HGNC:18420] | 33511 | -1.023 | -0.0953 | No |
| 46 | EGR2 | early growth response 2 [Source:HGNC Symbol;Acc:HGNC:3239] | 33799 | -1.111 | -0.0800 | No |
| 47 | PLCB3 | phospholipase C beta 3 [Source:HGNC Symbol;Acc:HGNC:9056] | 33927 | -1.159 | -0.0594 | No |
| 48 | SLC25A4 | solute carrier family 25 member 4 [Source:HGNC Symbol;Acc:HGNC:10990] | 34208 | -1.248 | -0.0411 | No |
| 49 | DIXDC1 | DIX domain containing 1 [Source:HGNC Symbol;Acc:HGNC:23695] | 35274 | -1.716 | -0.0344 | No |
| 50 | ANXA6 | annexin A6 [Source:HGNC Symbol;Acc:HGNC:544] | 35382 | -1.767 | -0.0006 | No |
| 51 | VCL | vinculin [Source:HGNC Symbol;Acc:HGNC:12665] | 36071 | -2.255 | 0.0275 | No |