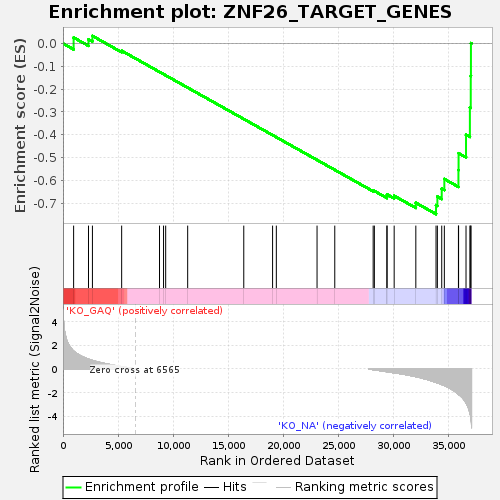
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | ZNF26_TARGET_GENES |
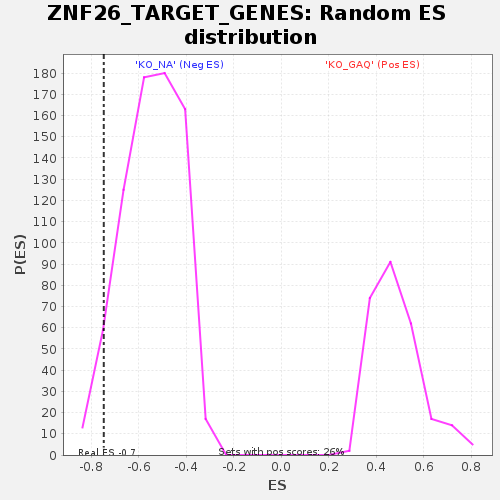
| Enrichment Score (ES) | -0.74731636 |
| Normalized Enrichment Score (NES) | -1.3712475 |
| Nominal p-value | 0.05170068 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SNX16 | sorting nexin 16 [Source:HGNC Symbol;Acc:HGNC:14980] | 964 | 1.505 | 0.0261 | No |
| 2 | LYPLA2P3 | LYPLA2 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:50446] | 2314 | 0.823 | 0.0182 | No |
| 3 | COX16 | cytochrome c oxidase assembly factor COX16 [Source:HGNC Symbol;Acc:HGNC:20213] | 2671 | 0.710 | 0.0331 | No |
| 4 | FABP5P7 | fatty acid binding protein 5 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:31070] | 5325 | 0.188 | -0.0319 | No |
| 5 | LINC01251 | long intergenic non-protein coding RNA 1251 [Source:HGNC Symbol;Acc:HGNC:49845] | 8761 | 0.000 | -0.1246 | No |
| 6 | LINC01119 | long intergenic non-protein coding RNA 1119 [Source:HGNC Symbol;Acc:HGNC:49262] | 9127 | 0.000 | -0.1345 | No |
| 7 | SATB1-AS1 | SATB1 antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:50687] | 9321 | 0.000 | -0.1397 | No |
| 8 | SRGAP1 | SLIT-ROBO Rho GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:17382] | 11318 | 0.000 | -0.1935 | No |
| 9 | ZNF561-AS1 | ZNF561 antisense RNA 1 (head to head) [Source:HGNC Symbol;Acc:HGNC:27613] | 16410 | 0.000 | -0.3309 | No |
| 10 | OSMR | oncostatin M receptor [Source:HGNC Symbol;Acc:HGNC:8507] | 19026 | 0.000 | -0.4015 | No |
| 11 | OTOS | otospiralin [Source:HGNC Symbol;Acc:HGNC:22644] | 19366 | 0.000 | -0.4106 | No |
| 12 | CCL8 | C-C motif chemokine ligand 8 [Source:HGNC Symbol;Acc:HGNC:10635] | 23062 | 0.000 | -0.5103 | No |
| 13 | LINC00431 | long intergenic non-protein coding RNA 431 [Source:HGNC Symbol;Acc:HGNC:42766] | 24671 | 0.000 | -0.5537 | No |
| 14 | TAGLN2P1 | transgelin 2 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:21739] | 28156 | -0.057 | -0.6457 | No |
| 15 | STAT6 | signal transducer and activator of transcription 6 [Source:HGNC Symbol;Acc:HGNC:11368] | 28266 | -0.073 | -0.6462 | No |
| 16 | ZNF567 | zinc finger protein 567 [Source:HGNC Symbol;Acc:HGNC:28696] | 29407 | -0.232 | -0.6689 | No |
| 17 | CLCN5 | chloride voltage-gated channel 5 [Source:HGNC Symbol;Acc:HGNC:2023] | 29414 | -0.233 | -0.6610 | No |
| 18 | ERICH1 | glutamate rich 1 [Source:HGNC Symbol;Acc:HGNC:27234] | 30063 | -0.325 | -0.6672 | No |
| 19 | WWP2 | WW domain containing E3 ubiquitin protein ligase 2 [Source:HGNC Symbol;Acc:HGNC:16804] | 32029 | -0.647 | -0.6978 | No |
| 20 | OSBPL9 | oxysterol binding protein like 9 [Source:HGNC Symbol;Acc:HGNC:16386] | 33864 | -1.134 | -0.7081 | Yes |
| 21 | UAP1 | UDP-N-acetylglucosamine pyrophosphorylase 1 [Source:HGNC Symbol;Acc:HGNC:12457] | 33986 | -1.181 | -0.6705 | Yes |
| 22 | ZNF45 | zinc finger protein 45 [Source:HGNC Symbol;Acc:HGNC:13111] | 34381 | -1.312 | -0.6357 | Yes |
| 23 | GSTO1 | glutathione S-transferase omega 1 [Source:HGNC Symbol;Acc:HGNC:13312] | 34618 | -1.399 | -0.5937 | Yes |
| 24 | UHRF1 | ubiquitin like with PHD and ring finger domains 1 [Source:HGNC Symbol;Acc:HGNC:12556] | 35897 | -2.126 | -0.5546 | Yes |
| 25 | MPPE1 | metallophosphoesterase 1 [Source:HGNC Symbol;Acc:HGNC:15988] | 35911 | -2.142 | -0.4808 | Yes |
| 26 | BBC3 | BCL2 binding component 3 [Source:HGNC Symbol;Acc:HGNC:17868] | 36586 | -2.867 | -0.3998 | Yes |
| 27 | E2F2 | E2F transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:3114] | 36937 | -3.731 | -0.2801 | Yes |
| 28 | PFKM | phosphofructokinase, muscle [Source:HGNC Symbol;Acc:HGNC:8877] | 37015 | -4.046 | -0.1422 | Yes |
| 29 | OMG | oligodendrocyte myelin glycoprotein [Source:HGNC Symbol;Acc:HGNC:8135] | 37032 | -4.166 | 0.0016 | Yes |