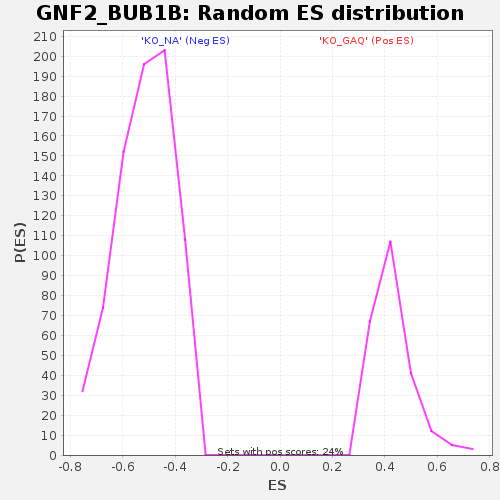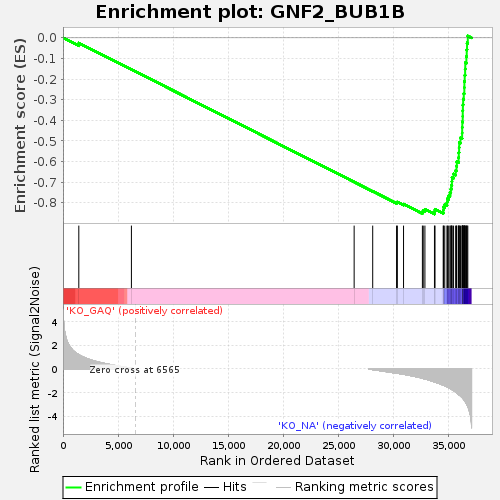
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_BUB1B |
| Enrichment Score (ES) | -0.85474926 |
| Normalized Enrichment Score (NES) | -1.6549029 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0011828404 |
| FWER p-Value | 0.015 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PA2G4 | proliferation-associated 2G4 [Source:HGNC Symbol;Acc:HGNC:8550] | 1435 | 1.187 | -0.0252 | No |
| 2 | MRPL39 | mitochondrial ribosomal protein L39 [Source:HGNC Symbol;Acc:HGNC:14027] | 6210 | 0.059 | -0.1535 | No |
| 3 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26430 | 0.000 | -0.6993 | No |
| 4 | TCP1 | t-complex 1 [Source:HGNC Symbol;Acc:HGNC:11655] | 28115 | -0.049 | -0.7442 | No |
| 5 | NUP205 | nucleoporin 205 [Source:HGNC Symbol;Acc:HGNC:18658] | 30281 | -0.352 | -0.7987 | No |
| 6 | HJURP | Holliday junction recognition protein [Source:HGNC Symbol;Acc:HGNC:25444] | 30352 | -0.363 | -0.7964 | No |
| 7 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 30914 | -0.450 | -0.8064 | No |
| 8 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 32625 | -0.776 | -0.8438 | Yes |
| 9 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32700 | -0.796 | -0.8367 | Yes |
| 10 | LMNB1 | lamin B1 [Source:HGNC Symbol;Acc:HGNC:6637] | 32854 | -0.836 | -0.8314 | Yes |
| 11 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.8424 | Yes |
| 12 | KIF20A | kinesin family member 20A [Source:HGNC Symbol;Acc:HGNC:9787] | 33771 | -1.101 | -0.8312 | Yes |
| 13 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34522 | -1.361 | -0.8360 | Yes |
| 14 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34533 | -1.364 | -0.8208 | Yes |
| 15 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34655 | -1.411 | -0.8080 | Yes |
| 16 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 34860 | -1.492 | -0.7966 | Yes |
| 17 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34896 | -1.510 | -0.7803 | Yes |
| 18 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35012 | -1.573 | -0.7656 | Yes |
| 19 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35131 | -1.642 | -0.7501 | Yes |
| 20 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 35207 | -1.677 | -0.7331 | Yes |
| 21 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 35266 | -1.714 | -0.7151 | Yes |
| 22 | ASF1B | anti-silencing function 1B histone chaperone [Source:HGNC Symbol;Acc:HGNC:20996] | 35303 | -1.732 | -0.6964 | Yes |
| 23 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35315 | -1.737 | -0.6770 | Yes |
| 24 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35455 | -1.811 | -0.6601 | Yes |
| 25 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35645 | -1.933 | -0.6433 | Yes |
| 26 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35723 | -1.996 | -0.6227 | Yes |
| 27 | HELLS | helicase, lymphoid specific [Source:HGNC Symbol;Acc:HGNC:4861] | 35750 | -2.017 | -0.6005 | Yes |
| 28 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35912 | -2.142 | -0.5805 | Yes |
| 29 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35930 | -2.153 | -0.5564 | Yes |
| 30 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35959 | -2.181 | -0.5324 | Yes |
| 31 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 35968 | -2.186 | -0.5078 | Yes |
| 32 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36081 | -2.263 | -0.4851 | Yes |
| 33 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36232 | -2.394 | -0.4619 | Yes |
| 34 | KIF14 | kinesin family member 14 [Source:HGNC Symbol;Acc:HGNC:19181] | 36238 | -2.399 | -0.4348 | Yes |
| 35 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1148] | 36249 | -2.411 | -0.4077 | Yes |
| 36 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36286 | -2.451 | -0.3808 | Yes |
| 37 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36289 | -2.458 | -0.3529 | Yes |
| 38 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36301 | -2.470 | -0.3251 | Yes |
| 39 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36326 | -2.499 | -0.2974 | Yes |
| 40 | CENPM | centromere protein M [Source:HGNC Symbol;Acc:HGNC:18352] | 36383 | -2.566 | -0.2697 | Yes |
| 41 | CEP55 | centrosomal protein 55 [Source:HGNC Symbol;Acc:HGNC:1161] | 36433 | -2.620 | -0.2413 | Yes |
| 42 | ESPL1 | extra spindle pole bodies like 1, separase [Source:HGNC Symbol;Acc:HGNC:16856] | 36438 | -2.629 | -0.2115 | Yes |
| 43 | PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase [Source:HGNC Symbol;Acc:HGNC:8587] | 36467 | -2.673 | -0.1819 | Yes |
| 44 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36505 | -2.720 | -0.1520 | Yes |
| 45 | NCAPD2 | non-SMC condensin I complex subunit D2 [Source:HGNC Symbol;Acc:HGNC:24305] | 36519 | -2.751 | -0.1211 | Yes |
| 46 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36620 | -2.903 | -0.0908 | Yes |
| 47 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36649 | -2.950 | -0.0580 | Yes |
| 48 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36679 | -3.000 | -0.0247 | Yes |
| 49 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36747 | -3.148 | 0.0093 | Yes |