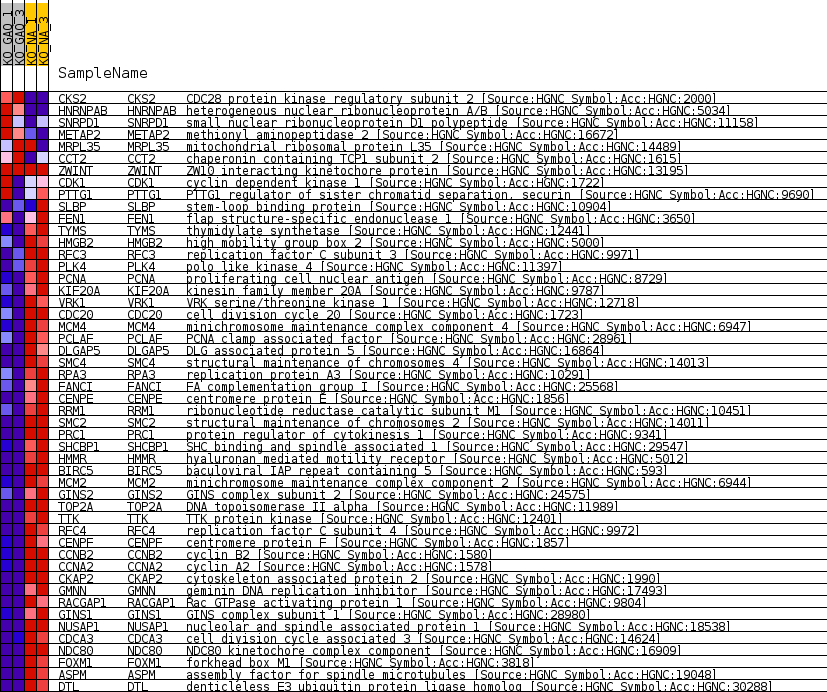
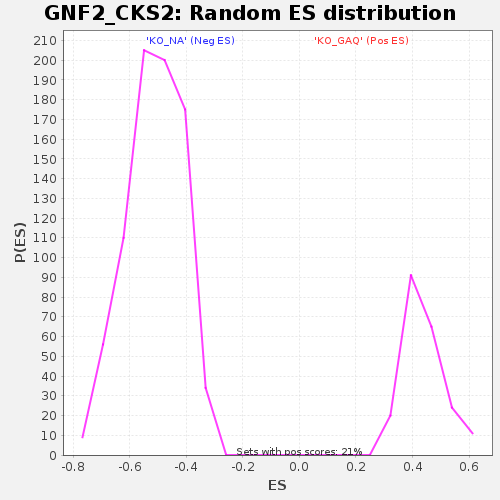
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_CKS2 |
| Enrichment Score (ES) | -0.84248066 |
| Normalized Enrichment Score (NES) | -1.6414189 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0014830756 |
| FWER p-Value | 0.024 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1504 | 1.154 | -0.0263 | No |
| 2 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B [Source:HGNC Symbol;Acc:HGNC:5034] | 2049 | 0.918 | -0.0296 | No |
| 3 | SNRPD1 | small nuclear ribonucleoprotein D1 polypeptide [Source:HGNC Symbol;Acc:HGNC:11158] | 4894 | 0.258 | -0.1032 | No |
| 4 | METAP2 | methionyl aminopeptidase 2 [Source:HGNC Symbol;Acc:HGNC:16672] | 5239 | 0.202 | -0.1100 | No |
| 5 | MRPL35 | mitochondrial ribosomal protein L35 [Source:HGNC Symbol;Acc:HGNC:14489] | 5592 | 0.147 | -0.1177 | No |
| 6 | CCT2 | chaperonin containing TCP1 subunit 2 [Source:HGNC Symbol;Acc:HGNC:1615] | 5629 | 0.142 | -0.1169 | No |
| 7 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26430 | 0.000 | -0.6784 | No |
| 8 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27912 | -0.021 | -0.7181 | No |
| 9 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 28646 | -0.123 | -0.7364 | No |
| 10 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 30070 | -0.326 | -0.7708 | No |
| 11 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 30452 | -0.375 | -0.7764 | No |
| 12 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32700 | -0.796 | -0.8272 | No |
| 13 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33267 | -0.949 | -0.8307 | Yes |
| 14 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33690 | -1.077 | -0.8288 | Yes |
| 15 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 33697 | -1.078 | -0.8156 | Yes |
| 16 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.8027 | Yes |
| 17 | KIF20A | kinesin family member 20A [Source:HGNC Symbol;Acc:HGNC:9787] | 33771 | -1.101 | -0.7904 | Yes |
| 18 | VRK1 | VRK serine/threonine kinase 1 [Source:HGNC Symbol;Acc:HGNC:12718] | 34454 | -1.336 | -0.7923 | Yes |
| 19 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34533 | -1.364 | -0.7775 | Yes |
| 20 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34655 | -1.411 | -0.7632 | Yes |
| 21 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34665 | -1.416 | -0.7459 | Yes |
| 22 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 34894 | -1.508 | -0.7334 | Yes |
| 23 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34896 | -1.510 | -0.7147 | Yes |
| 24 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 34976 | -1.554 | -0.6976 | Yes |
| 25 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35012 | -1.573 | -0.6790 | Yes |
| 26 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 35165 | -1.659 | -0.6626 | Yes |
| 27 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35455 | -1.811 | -0.6479 | Yes |
| 28 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35493 | -1.831 | -0.6262 | Yes |
| 29 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 35527 | -1.854 | -0.6041 | Yes |
| 30 | SHCBP1 | SHC binding and spindle associated 1 [Source:HGNC Symbol;Acc:HGNC:29547] | 35572 | -1.882 | -0.5820 | Yes |
| 31 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35723 | -1.996 | -0.5613 | Yes |
| 32 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 35889 | -2.122 | -0.5394 | Yes |
| 33 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35912 | -2.142 | -0.5135 | Yes |
| 34 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35922 | -2.150 | -0.4871 | Yes |
| 35 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35959 | -2.181 | -0.4610 | Yes |
| 36 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 35968 | -2.186 | -0.4341 | Yes |
| 37 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36015 | -2.212 | -0.4080 | Yes |
| 38 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36049 | -2.239 | -0.3811 | Yes |
| 39 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36081 | -2.263 | -0.3539 | Yes |
| 40 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36232 | -2.394 | -0.3283 | Yes |
| 41 | CKAP2 | cytoskeleton associated protein 2 [Source:HGNC Symbol;Acc:HGNC:1990] | 36277 | -2.444 | -0.2992 | Yes |
| 42 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36301 | -2.470 | -0.2692 | Yes |
| 43 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36326 | -2.499 | -0.2388 | Yes |
| 44 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36505 | -2.720 | -0.2099 | Yes |
| 45 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36555 | -2.800 | -0.1766 | Yes |
| 46 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36562 | -2.813 | -0.1419 | Yes |
| 47 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36620 | -2.903 | -0.1074 | Yes |
| 48 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36696 | -3.045 | -0.0717 | Yes |
| 49 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36747 | -3.148 | -0.0340 | Yes |
| 50 | DTL | denticleless E3 ubiquitin protein ligase homolog [Source:HGNC Symbol;Acc:HGNC:30288] | 36872 | -3.490 | 0.0059 | Yes |