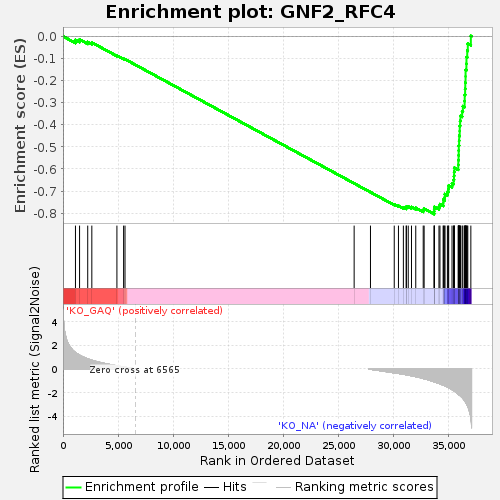
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GNF2_RFC4 |
| Enrichment Score (ES) | -0.8039972 |
| Normalized Enrichment Score (NES) | -1.6127537 |
| Nominal p-value | 0.0012738854 |
| FDR q-value | 0.0033051795 |
| FWER p-Value | 0.067 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DKC1 | dyskerin pseudouridine synthase 1 [Source:HGNC Symbol;Acc:HGNC:2890] | 1131 | 1.374 | -0.0163 | No |
| 2 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1504 | 1.154 | -0.0144 | No |
| 3 | RANBP1 | RAN binding protein 1 [Source:HGNC Symbol;Acc:HGNC:9847] | 2244 | 0.846 | -0.0256 | No |
| 4 | RAN | RAN, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9846] | 2617 | 0.727 | -0.0281 | No |
| 5 | SNRPD1 | small nuclear ribonucleoprotein D1 polypeptide [Source:HGNC Symbol;Acc:HGNC:11158] | 4894 | 0.258 | -0.0869 | No |
| 6 | SNRPE | small nuclear ribonucleoprotein polypeptide E [Source:HGNC Symbol;Acc:HGNC:11161] | 5502 | 0.162 | -0.1016 | No |
| 7 | CCT2 | chaperonin containing TCP1 subunit 2 [Source:HGNC Symbol;Acc:HGNC:1615] | 5629 | 0.142 | -0.1035 | No |
| 8 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26430 | 0.000 | -0.6652 | No |
| 9 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27912 | -0.021 | -0.7050 | No |
| 10 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 30070 | -0.326 | -0.7598 | No |
| 11 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 30452 | -0.375 | -0.7662 | No |
| 12 | SSRP1 | structure specific recognition protein 1 [Source:HGNC Symbol;Acc:HGNC:11327] | 30899 | -0.449 | -0.7736 | No |
| 13 | MCM10 | minichromosome maintenance 10 replication initiation factor [Source:HGNC Symbol;Acc:HGNC:18043] | 31158 | -0.486 | -0.7756 | No |
| 14 | MLH1 | mutL homolog 1 [Source:HGNC Symbol;Acc:HGNC:7127] | 31170 | -0.489 | -0.7708 | No |
| 15 | CTPS1 | CTP synthase 1 [Source:HGNC Symbol;Acc:HGNC:2519] | 31341 | -0.517 | -0.7700 | No |
| 16 | ITGB3BP | integrin subunit beta 3 binding protein [Source:HGNC Symbol;Acc:HGNC:6157] | 31636 | -0.567 | -0.7721 | No |
| 17 | SUPT16H | SPT16 homolog, facilitates chromatin remodeling subunit [Source:HGNC Symbol;Acc:HGNC:11465] | 32023 | -0.646 | -0.7758 | No |
| 18 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32700 | -0.796 | -0.7858 | No |
| 19 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 32783 | -0.819 | -0.7795 | No |
| 20 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33690 | -1.077 | -0.7928 | Yes |
| 21 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 33697 | -1.078 | -0.7818 | Yes |
| 22 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.7712 | Yes |
| 23 | TIMELESS | timeless circadian regulator [Source:HGNC Symbol;Acc:HGNC:11813] | 34130 | -1.218 | -0.7696 | Yes |
| 24 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34219 | -1.252 | -0.7590 | Yes |
| 25 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34522 | -1.361 | -0.7530 | Yes |
| 26 | KNTC1 | kinetochore associated 1 [Source:HGNC Symbol;Acc:HGNC:17255] | 34524 | -1.362 | -0.7389 | Yes |
| 27 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34655 | -1.411 | -0.7278 | Yes |
| 28 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34665 | -1.416 | -0.7134 | Yes |
| 29 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34896 | -1.510 | -0.7039 | Yes |
| 30 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 34976 | -1.554 | -0.6900 | Yes |
| 31 | FANCI | FA complementation group I [Source:HGNC Symbol;Acc:HGNC:25568] | 35012 | -1.573 | -0.6746 | Yes |
| 32 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 35315 | -1.737 | -0.6647 | Yes |
| 33 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 35455 | -1.811 | -0.6497 | Yes |
| 34 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35493 | -1.831 | -0.6317 | Yes |
| 35 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35508 | -1.842 | -0.6130 | Yes |
| 36 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 35534 | -1.859 | -0.5944 | Yes |
| 37 | RAD54L | RAD54 like [Source:HGNC Symbol;Acc:HGNC:9826] | 35882 | -2.115 | -0.5819 | Yes |
| 38 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 35889 | -2.122 | -0.5600 | Yes |
| 39 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35912 | -2.142 | -0.5384 | Yes |
| 40 | GINS2 | GINS complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:24575] | 35922 | -2.150 | -0.5164 | Yes |
| 41 | KIF11 | kinesin family member 11 [Source:HGNC Symbol;Acc:HGNC:6388] | 35930 | -2.153 | -0.4942 | Yes |
| 42 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35959 | -2.181 | -0.4724 | Yes |
| 43 | TTK | TTK protein kinase [Source:HGNC Symbol;Acc:HGNC:12401] | 35968 | -2.186 | -0.4499 | Yes |
| 44 | MCM6 | minichromosome maintenance complex component 6 [Source:HGNC Symbol;Acc:HGNC:6949] | 36012 | -2.211 | -0.4282 | Yes |
| 45 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36015 | -2.212 | -0.4053 | Yes |
| 46 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36049 | -2.239 | -0.3830 | Yes |
| 47 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36081 | -2.263 | -0.3604 | Yes |
| 48 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36232 | -2.394 | -0.3396 | Yes |
| 49 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 36301 | -2.470 | -0.3158 | Yes |
| 50 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 36458 | -2.656 | -0.2925 | Yes |
| 51 | PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase [Source:HGNC Symbol;Acc:HGNC:8587] | 36467 | -2.673 | -0.2650 | Yes |
| 52 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36505 | -2.720 | -0.2378 | Yes |
| 53 | NCAPD2 | non-SMC condensin I complex subunit D2 [Source:HGNC Symbol;Acc:HGNC:24305] | 36519 | -2.751 | -0.2096 | Yes |
| 54 | MSH6 | mutS homolog 6 [Source:HGNC Symbol;Acc:HGNC:7329] | 36544 | -2.786 | -0.1814 | Yes |
| 55 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36546 | -2.789 | -0.1525 | Yes |
| 56 | NDC80 | NDC80 kinetochore complex component [Source:HGNC Symbol;Acc:HGNC:16909] | 36620 | -2.903 | -0.1244 | Yes |
| 57 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36627 | -2.912 | -0.0944 | Yes |
| 58 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36696 | -3.045 | -0.0646 | Yes |
| 59 | ASPM | assembly factor for spindle microtubules [Source:HGNC Symbol;Acc:HGNC:19048] | 36747 | -3.148 | -0.0334 | Yes |
| 60 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 37024 | -4.108 | 0.0018 | Yes |

