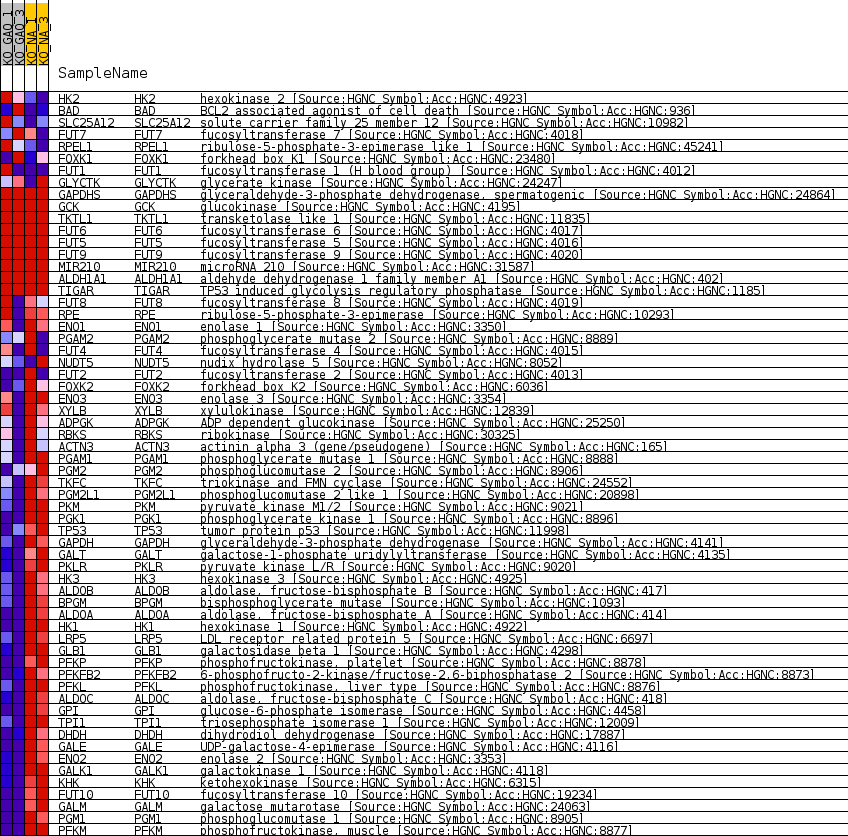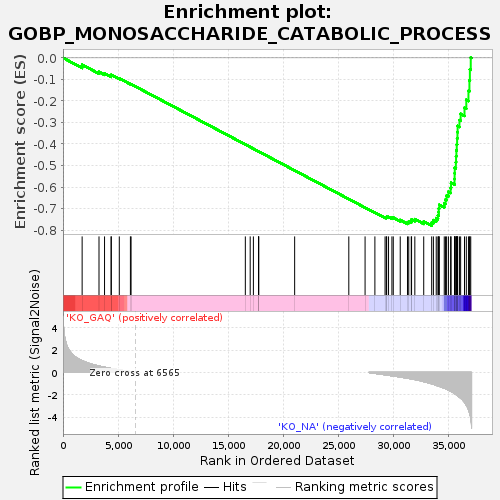
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | GOBP_MONOSACCHARIDE_CATABOLIC_PROCESS |
| Enrichment Score (ES) | -0.77890694 |
| Normalized Enrichment Score (NES) | -1.5823221 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.1050854 |
| FWER p-Value | 0.884 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HK2 | hexokinase 2 [Source:HGNC Symbol;Acc:HGNC:4923] | 1737 | 1.045 | -0.0321 | No |
| 2 | BAD | BCL2 associated agonist of cell death [Source:HGNC Symbol;Acc:HGNC:936] | 3268 | 0.567 | -0.0653 | No |
| 3 | SLC25A12 | solute carrier family 25 member 12 [Source:HGNC Symbol;Acc:HGNC:10982] | 3771 | 0.458 | -0.0723 | No |
| 4 | FUT7 | fucosyltransferase 7 [Source:HGNC Symbol;Acc:HGNC:4018] | 4366 | 0.349 | -0.0834 | No |
| 5 | RPEL1 | ribulose-5-phosphate-3-epimerase like 1 [Source:HGNC Symbol;Acc:HGNC:45241] | 4384 | 0.346 | -0.0790 | No |
| 6 | FOXK1 | forkhead box K1 [Source:HGNC Symbol;Acc:HGNC:23480] | 5110 | 0.221 | -0.0954 | No |
| 7 | FUT1 | fucosyltransferase 1 (H blood group) [Source:HGNC Symbol;Acc:HGNC:4012] | 6119 | 0.075 | -0.1215 | No |
| 8 | GLYCTK | glycerate kinase [Source:HGNC Symbol;Acc:HGNC:24247] | 6179 | 0.065 | -0.1222 | No |
| 9 | GAPDHS | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic [Source:HGNC Symbol;Acc:HGNC:24864] | 16552 | 0.000 | -0.4023 | No |
| 10 | GCK | glucokinase [Source:HGNC Symbol;Acc:HGNC:4195] | 16993 | 0.000 | -0.4142 | No |
| 11 | TKTL1 | transketolase like 1 [Source:HGNC Symbol;Acc:HGNC:11835] | 17283 | 0.000 | -0.4220 | No |
| 12 | FUT6 | fucosyltransferase 6 [Source:HGNC Symbol;Acc:HGNC:4017] | 17757 | 0.000 | -0.4348 | No |
| 13 | FUT5 | fucosyltransferase 5 [Source:HGNC Symbol;Acc:HGNC:4016] | 17758 | 0.000 | -0.4348 | No |
| 14 | FUT9 | fucosyltransferase 9 [Source:HGNC Symbol;Acc:HGNC:4020] | 17759 | 0.000 | -0.4348 | No |
| 15 | MIR210 | microRNA 210 [Source:HGNC Symbol;Acc:HGNC:31587] | 21029 | 0.000 | -0.5231 | No |
| 16 | ALDH1A1 | aldehyde dehydrogenase 1 family member A1 [Source:HGNC Symbol;Acc:HGNC:402] | 25945 | 0.000 | -0.6558 | No |
| 17 | TIGAR | TP53 induced glycolysis regulatory phosphatase [Source:HGNC Symbol;Acc:HGNC:1185] | 27422 | 0.000 | -0.6956 | No |
| 18 | FUT8 | fucosyltransferase 8 [Source:HGNC Symbol;Acc:HGNC:4019] | 28313 | -0.079 | -0.7186 | No |
| 19 | RPE | ribulose-5-phosphate-3-epimerase [Source:HGNC Symbol;Acc:HGNC:10293] | 29237 | -0.206 | -0.7406 | No |
| 20 | ENO1 | enolase 1 [Source:HGNC Symbol;Acc:HGNC:3350] | 29354 | -0.225 | -0.7405 | No |
| 21 | PGAM2 | phosphoglycerate mutase 2 [Source:HGNC Symbol;Acc:HGNC:8889] | 29383 | -0.229 | -0.7380 | No |
| 22 | FUT4 | fucosyltransferase 4 [Source:HGNC Symbol;Acc:HGNC:4015] | 29552 | -0.250 | -0.7390 | No |
| 23 | NUDT5 | nudix hydrolase 5 [Source:HGNC Symbol;Acc:HGNC:8052] | 29859 | -0.297 | -0.7430 | No |
| 24 | FUT2 | fucosyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:4013] | 29982 | -0.314 | -0.7419 | No |
| 25 | FOXK2 | forkhead box K2 [Source:HGNC Symbol;Acc:HGNC:6036] | 30614 | -0.404 | -0.7532 | No |
| 26 | ENO3 | enolase 3 [Source:HGNC Symbol;Acc:HGNC:3354] | 31272 | -0.506 | -0.7637 | No |
| 27 | XYLB | xylulokinase [Source:HGNC Symbol;Acc:HGNC:12839] | 31387 | -0.525 | -0.7593 | No |
| 28 | ADPGK | ADP dependent glucokinase [Source:HGNC Symbol;Acc:HGNC:25250] | 31615 | -0.564 | -0.7574 | No |
| 29 | RBKS | ribokinase [Source:HGNC Symbol;Acc:HGNC:30325] | 31643 | -0.568 | -0.7501 | No |
| 30 | ACTN3 | actinin alpha 3 (gene/pseudogene) [Source:HGNC Symbol;Acc:HGNC:165] | 31939 | -0.626 | -0.7492 | No |
| 31 | PGAM1 | phosphoglycerate mutase 1 [Source:HGNC Symbol;Acc:HGNC:8888] | 32746 | -0.810 | -0.7594 | No |
| 32 | PGM2 | phosphoglucomutase 2 [Source:HGNC Symbol;Acc:HGNC:8906] | 33469 | -1.010 | -0.7645 | Yes |
| 33 | TKFC | triokinase and FMN cyclase [Source:HGNC Symbol;Acc:HGNC:24552] | 33624 | -1.055 | -0.7537 | Yes |
| 34 | PGM2L1 | phosphoglucomutase 2 like 1 [Source:HGNC Symbol;Acc:HGNC:20898] | 33882 | -1.139 | -0.7444 | Yes |
| 35 | PKM | pyruvate kinase M1/2 [Source:HGNC Symbol;Acc:HGNC:9021] | 34036 | -1.195 | -0.7316 | Yes |
| 36 | PGK1 | phosphoglycerate kinase 1 [Source:HGNC Symbol;Acc:HGNC:8896] | 34100 | -1.209 | -0.7161 | Yes |
| 37 | TP53 | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 34112 | -1.214 | -0.6991 | Yes |
| 38 | GAPDH | glyceraldehyde-3-phosphate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4141] | 34158 | -1.229 | -0.6828 | Yes |
| 39 | GALT | galactose-1-phosphate uridylyltransferase [Source:HGNC Symbol;Acc:HGNC:4135] | 34610 | -1.396 | -0.6752 | Yes |
| 40 | PKLR | pyruvate kinase L/R [Source:HGNC Symbol;Acc:HGNC:9020] | 34728 | -1.436 | -0.6579 | Yes |
| 41 | HK3 | hexokinase 3 [Source:HGNC Symbol;Acc:HGNC:4925] | 34823 | -1.473 | -0.6395 | Yes |
| 42 | ALDOB | aldolase, fructose-bisphosphate B [Source:HGNC Symbol;Acc:HGNC:417] | 34988 | -1.560 | -0.6217 | Yes |
| 43 | BPGM | bisphosphoglycerate mutase [Source:HGNC Symbol;Acc:HGNC:1093] | 35189 | -1.666 | -0.6034 | Yes |
| 44 | ALDOA | aldolase, fructose-bisphosphate A [Source:HGNC Symbol;Acc:HGNC:414] | 35228 | -1.689 | -0.5804 | Yes |
| 45 | HK1 | hexokinase 1 [Source:HGNC Symbol;Acc:HGNC:4922] | 35541 | -1.862 | -0.5624 | Yes |
| 46 | LRP5 | LDL receptor related protein 5 [Source:HGNC Symbol;Acc:HGNC:6697] | 35551 | -1.867 | -0.5361 | Yes |
| 47 | GLB1 | galactosidase beta 1 [Source:HGNC Symbol;Acc:HGNC:4298] | 35563 | -1.874 | -0.5097 | Yes |
| 48 | PFKP | phosphofructokinase, platelet [Source:HGNC Symbol;Acc:HGNC:8878] | 35663 | -1.948 | -0.4847 | Yes |
| 49 | PFKFB2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 [Source:HGNC Symbol;Acc:HGNC:8873] | 35682 | -1.962 | -0.4573 | Yes |
| 50 | PFKL | phosphofructokinase, liver type [Source:HGNC Symbol;Acc:HGNC:8876] | 35711 | -1.983 | -0.4298 | Yes |
| 51 | ALDOC | aldolase, fructose-bisphosphate C [Source:HGNC Symbol;Acc:HGNC:418] | 35745 | -2.014 | -0.4021 | Yes |
| 52 | GPI | glucose-6-phosphate isomerase [Source:HGNC Symbol;Acc:HGNC:4458] | 35778 | -2.043 | -0.3739 | Yes |
| 53 | TPI1 | triosephosphate isomerase 1 [Source:HGNC Symbol;Acc:HGNC:12009] | 35808 | -2.065 | -0.3453 | Yes |
| 54 | DHDH | dihydrodiol dehydrogenase [Source:HGNC Symbol;Acc:HGNC:17887] | 35824 | -2.072 | -0.3162 | Yes |
| 55 | GALE | UDP-galactose-4-epimerase [Source:HGNC Symbol;Acc:HGNC:4116] | 35988 | -2.197 | -0.2894 | Yes |
| 56 | ENO2 | enolase 2 [Source:HGNC Symbol;Acc:HGNC:3353] | 36098 | -2.276 | -0.2600 | Yes |
| 57 | GALK1 | galactokinase 1 [Source:HGNC Symbol;Acc:HGNC:4118] | 36448 | -2.645 | -0.2318 | Yes |
| 58 | KHK | ketohexokinase [Source:HGNC Symbol;Acc:HGNC:6315] | 36612 | -2.893 | -0.1950 | Yes |
| 59 | FUT10 | fucosyltransferase 10 [Source:HGNC Symbol;Acc:HGNC:19234] | 36807 | -3.288 | -0.1535 | Yes |
| 60 | GALM | galactose mutarotase [Source:HGNC Symbol;Acc:HGNC:24063] | 36895 | -3.560 | -0.1052 | Yes |
| 61 | PGM1 | phosphoglucomutase 1 [Source:HGNC Symbol;Acc:HGNC:8905] | 36936 | -3.719 | -0.0534 | Yes |
| 62 | PFKM | phosphofructokinase, muscle [Source:HGNC Symbol;Acc:HGNC:8877] | 37015 | -4.046 | 0.0020 | Yes |