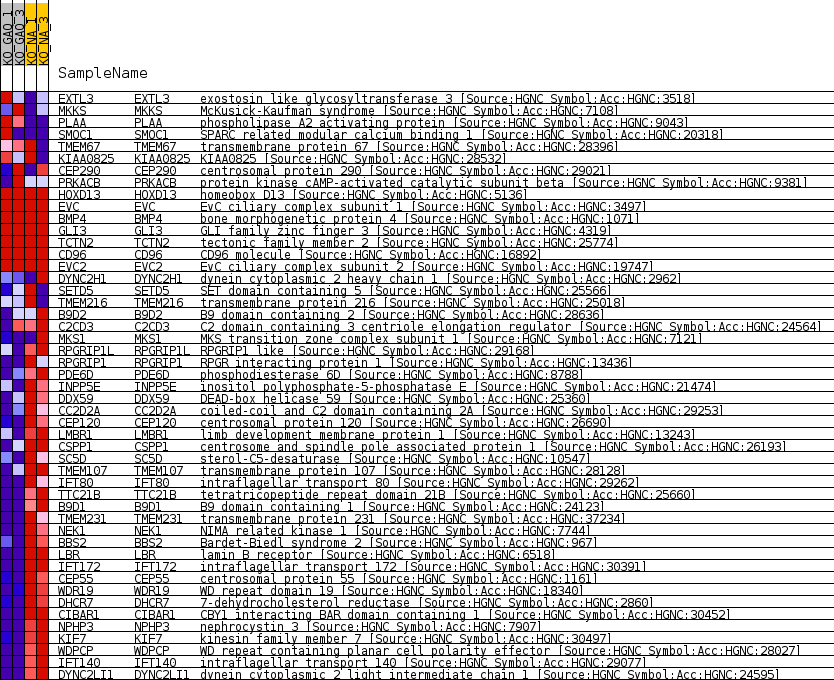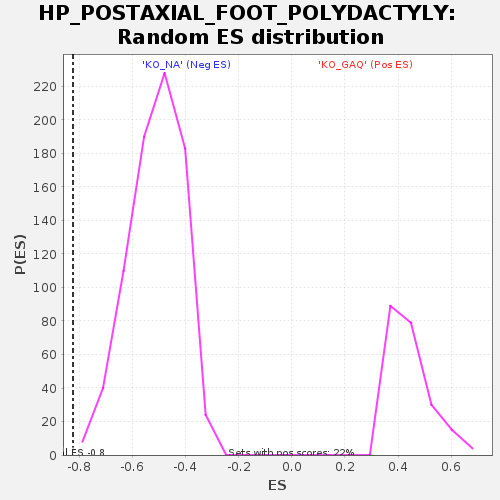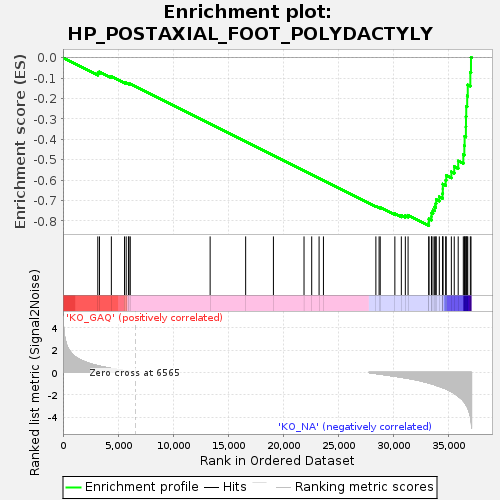
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HP_POSTAXIAL_FOOT_POLYDACTYLY |
| Enrichment Score (ES) | -0.82310444 |
| Normalized Enrichment Score (NES) | -1.6077033 |
| Nominal p-value | 0.0012771392 |
| FDR q-value | 0.065354474 |
| FWER p-Value | 0.658 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EXTL3 | exostosin like glycosyltransferase 3 [Source:HGNC Symbol;Acc:HGNC:3518] | 3159 | 0.593 | -0.0749 | No |
| 2 | MKKS | McKusick-Kaufman syndrome [Source:HGNC Symbol;Acc:HGNC:7108] | 3310 | 0.559 | -0.0691 | No |
| 3 | PLAA | phospholipase A2 activating protein [Source:HGNC Symbol;Acc:HGNC:9043] | 4390 | 0.346 | -0.0921 | No |
| 4 | SMOC1 | SPARC related modular calcium binding 1 [Source:HGNC Symbol;Acc:HGNC:20318] | 5585 | 0.147 | -0.1218 | No |
| 5 | TMEM67 | transmembrane protein 67 [Source:HGNC Symbol;Acc:HGNC:28396] | 5736 | 0.132 | -0.1235 | No |
| 6 | KIAA0825 | KIAA0825 [Source:HGNC Symbol;Acc:HGNC:28532] | 5940 | 0.099 | -0.1273 | No |
| 7 | CEP290 | centrosomal protein 290 [Source:HGNC Symbol;Acc:HGNC:29021] | 5991 | 0.090 | -0.1270 | No |
| 8 | PRKACB | protein kinase cAMP-activated catalytic subunit beta [Source:HGNC Symbol;Acc:HGNC:9381] | 6107 | 0.078 | -0.1288 | No |
| 9 | HOXD13 | homeobox D13 [Source:HGNC Symbol;Acc:HGNC:5136] | 13357 | 0.000 | -0.3245 | No |
| 10 | EVC | EvC ciliary complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:3497] | 16581 | 0.000 | -0.4115 | No |
| 11 | BMP4 | bone morphogenetic protein 4 [Source:HGNC Symbol;Acc:HGNC:1071] | 19102 | 0.000 | -0.4795 | No |
| 12 | GLI3 | GLI family zinc finger 3 [Source:HGNC Symbol;Acc:HGNC:4319] | 21880 | 0.000 | -0.5545 | No |
| 13 | TCTN2 | tectonic family member 2 [Source:HGNC Symbol;Acc:HGNC:25774] | 22577 | 0.000 | -0.5733 | No |
| 14 | CD96 | CD96 molecule [Source:HGNC Symbol;Acc:HGNC:16892] | 23247 | 0.000 | -0.5913 | No |
| 15 | EVC2 | EvC ciliary complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:19747] | 23643 | 0.000 | -0.6020 | No |
| 16 | DYNC2H1 | dynein cytoplasmic 2 heavy chain 1 [Source:HGNC Symbol;Acc:HGNC:2962] | 28402 | -0.089 | -0.7289 | No |
| 17 | SETD5 | SET domain containing 5 [Source:HGNC Symbol;Acc:HGNC:25566] | 28695 | -0.131 | -0.7344 | No |
| 18 | TMEM216 | transmembrane protein 216 [Source:HGNC Symbol;Acc:HGNC:25018] | 28797 | -0.143 | -0.7346 | No |
| 19 | B9D2 | B9 domain containing 2 [Source:HGNC Symbol;Acc:HGNC:28636] | 30126 | -0.333 | -0.7646 | No |
| 20 | C2CD3 | C2 domain containing 3 centriole elongation regulator [Source:HGNC Symbol;Acc:HGNC:24564] | 30714 | -0.418 | -0.7731 | No |
| 21 | MKS1 | MKS transition zone complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:7121] | 31077 | -0.473 | -0.7746 | No |
| 22 | RPGRIP1L | RPGRIP1 like [Source:HGNC Symbol;Acc:HGNC:29168] | 31326 | -0.515 | -0.7722 | No |
| 23 | RPGRIP1 | RPGR interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:13436] | 33211 | -0.934 | -0.8067 | Yes |
| 24 | PDE6D | phosphodiesterase 6D [Source:HGNC Symbol;Acc:HGNC:8788] | 33214 | -0.935 | -0.7903 | Yes |
| 25 | INPP5E | inositol polyphosphate-5-phosphatase E [Source:HGNC Symbol;Acc:HGNC:21474] | 33448 | -1.003 | -0.7790 | Yes |
| 26 | DDX59 | DEAD-box helicase 59 [Source:HGNC Symbol;Acc:HGNC:25360] | 33452 | -1.004 | -0.7614 | Yes |
| 27 | CC2D2A | coiled-coil and C2 domain containing 2A [Source:HGNC Symbol;Acc:HGNC:29253] | 33588 | -1.043 | -0.7467 | Yes |
| 28 | CEP120 | centrosomal protein 120 [Source:HGNC Symbol;Acc:HGNC:26690] | 33739 | -1.089 | -0.7317 | Yes |
| 29 | LMBR1 | limb development membrane protein 1 [Source:HGNC Symbol;Acc:HGNC:13243] | 33836 | -1.126 | -0.7145 | Yes |
| 30 | CSPP1 | centrosome and spindle pole associated protein 1 [Source:HGNC Symbol;Acc:HGNC:26193] | 33879 | -1.139 | -0.6956 | Yes |
| 31 | SC5D | sterol-C5-desaturase [Source:HGNC Symbol;Acc:HGNC:10547] | 34161 | -1.231 | -0.6816 | Yes |
| 32 | TMEM107 | transmembrane protein 107 [Source:HGNC Symbol;Acc:HGNC:28128] | 34460 | -1.339 | -0.6661 | Yes |
| 33 | IFT80 | intraflagellar transport 80 [Source:HGNC Symbol;Acc:HGNC:29262] | 34479 | -1.344 | -0.6430 | Yes |
| 34 | TTC21B | tetratricopeptide repeat domain 21B [Source:HGNC Symbol;Acc:HGNC:25660] | 34481 | -1.345 | -0.6194 | Yes |
| 35 | B9D1 | B9 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:24123] | 34738 | -1.439 | -0.6010 | Yes |
| 36 | TMEM231 | transmembrane protein 231 [Source:HGNC Symbol;Acc:HGNC:37234] | 34788 | -1.456 | -0.5767 | Yes |
| 37 | NEK1 | NIMA related kinase 1 [Source:HGNC Symbol;Acc:HGNC:7744] | 35255 | -1.707 | -0.5593 | Yes |
| 38 | BBS2 | Bardet-Biedl syndrome 2 [Source:HGNC Symbol;Acc:HGNC:967] | 35517 | -1.846 | -0.5339 | Yes |
| 39 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 35876 | -2.113 | -0.5065 | Yes |
| 40 | IFT172 | intraflagellar transport 172 [Source:HGNC Symbol;Acc:HGNC:30391] | 36340 | -2.513 | -0.4748 | Yes |
| 41 | CEP55 | centrosomal protein 55 [Source:HGNC Symbol;Acc:HGNC:1161] | 36433 | -2.620 | -0.4313 | Yes |
| 42 | WDR19 | WD repeat domain 19 [Source:HGNC Symbol;Acc:HGNC:18340] | 36459 | -2.656 | -0.3853 | Yes |
| 43 | DHCR7 | 7-dehydrocholesterol reductase [Source:HGNC Symbol;Acc:HGNC:2860] | 36576 | -2.839 | -0.3385 | Yes |
| 44 | CIBAR1 | CBY1 interacting BAR domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30452] | 36584 | -2.862 | -0.2885 | Yes |
| 45 | NPHP3 | nephrocystin 3 [Source:HGNC Symbol;Acc:HGNC:7907] | 36606 | -2.886 | -0.2383 | Yes |
| 46 | KIF7 | kinesin family member 7 [Source:HGNC Symbol;Acc:HGNC:30497] | 36692 | -3.041 | -0.1872 | Yes |
| 47 | WDPCP | WD repeat containing planar cell polarity effector [Source:HGNC Symbol;Acc:HGNC:28027] | 36744 | -3.144 | -0.1333 | Yes |
| 48 | IFT140 | intraflagellar transport 140 [Source:HGNC Symbol;Acc:HGNC:29077] | 36977 | -3.857 | -0.0718 | Yes |
| 49 | DYNC2LI1 | dynein cytoplasmic 2 light intermediate chain 1 [Source:HGNC Symbol;Acc:HGNC:24595] | 37037 | -4.261 | 0.0014 | Yes |