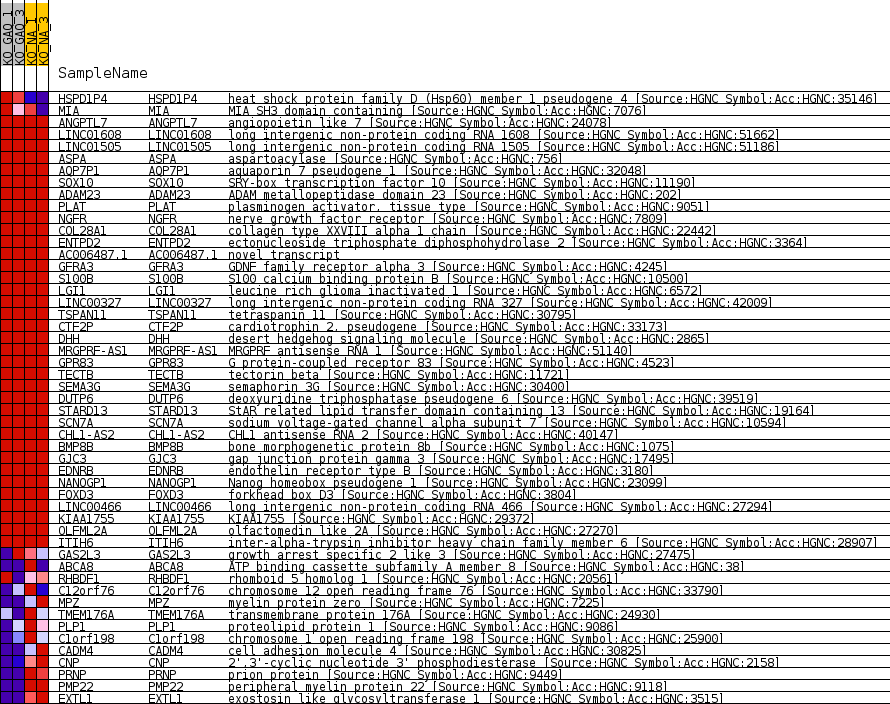
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | DESCARTES_MAIN_FETAL_SCHWANN_CELLS |
| Enrichment Score (ES) | -0.7715333 |
| Normalized Enrichment Score (NES) | -1.5091367 |
| Nominal p-value | 0.0038560412 |
| FDR q-value | 0.023947863 |
| FWER p-Value | 0.463 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | HSPD1P4 | heat shock protein family D (Hsp60) member 1 pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:35146] | 3076 | 0.615 | -0.0462 | No |
| 2 | MIA | MIA SH3 domain containing [Source:HGNC Symbol;Acc:HGNC:7076] | 3992 | 0.416 | -0.0459 | No |
| 3 | ANGPTL7 | angiopoietin like 7 [Source:HGNC Symbol;Acc:HGNC:24078] | 7185 | 0.000 | -0.1321 | No |
| 4 | LINC01608 | long intergenic non-protein coding RNA 1608 [Source:HGNC Symbol;Acc:HGNC:51662] | 7543 | 0.000 | -0.1418 | No |
| 5 | LINC01505 | long intergenic non-protein coding RNA 1505 [Source:HGNC Symbol;Acc:HGNC:51186] | 7815 | 0.000 | -0.1491 | No |
| 6 | ASPA | aspartoacylase [Source:HGNC Symbol;Acc:HGNC:756] | 7999 | 0.000 | -0.1540 | No |
| 7 | AQP7P1 | aquaporin 7 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:32048] | 8559 | 0.000 | -0.1691 | No |
| 8 | SOX10 | SRY-box transcription factor 10 [Source:HGNC Symbol;Acc:HGNC:11190] | 9010 | 0.000 | -0.1812 | No |
| 9 | ADAM23 | ADAM metallopeptidase domain 23 [Source:HGNC Symbol;Acc:HGNC:202] | 9899 | 0.000 | -0.2052 | No |
| 10 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] | 10092 | 0.000 | -0.2104 | No |
| 11 | NGFR | nerve growth factor receptor [Source:HGNC Symbol;Acc:HGNC:7809] | 10507 | 0.000 | -0.2216 | No |
| 12 | COL28A1 | collagen type XXVIII alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:22442] | 10558 | 0.000 | -0.2229 | No |
| 13 | ENTPD2 | ectonucleoside triphosphate diphosphohydrolase 2 [Source:HGNC Symbol;Acc:HGNC:3364] | 10599 | 0.000 | -0.2240 | No |
| 14 | AC006487.1 | novel transcript | 11348 | 0.000 | -0.2442 | No |
| 15 | GFRA3 | GDNF family receptor alpha 3 [Source:HGNC Symbol;Acc:HGNC:4245] | 11385 | 0.000 | -0.2452 | No |
| 16 | S100B | S100 calcium binding protein B [Source:HGNC Symbol;Acc:HGNC:10500] | 11806 | 0.000 | -0.2565 | No |
| 17 | LGI1 | leucine rich glioma inactivated 1 [Source:HGNC Symbol;Acc:HGNC:6572] | 12163 | 0.000 | -0.2661 | No |
| 18 | LINC00327 | long intergenic non-protein coding RNA 327 [Source:HGNC Symbol;Acc:HGNC:42009] | 13165 | 0.000 | -0.2932 | No |
| 19 | TSPAN11 | tetraspanin 11 [Source:HGNC Symbol;Acc:HGNC:30795] | 13811 | 0.000 | -0.3106 | No |
| 20 | CTF2P | cardiotrophin 2, pseudogene [Source:HGNC Symbol;Acc:HGNC:33173] | 15388 | 0.000 | -0.3531 | No |
| 21 | DHH | desert hedgehog signaling molecule [Source:HGNC Symbol;Acc:HGNC:2865] | 16166 | 0.000 | -0.3741 | No |
| 22 | MRGPRF-AS1 | MRGPRF antisense RNA 1 [Source:HGNC Symbol;Acc:HGNC:51140] | 16958 | 0.000 | -0.3954 | No |
| 23 | GPR83 | G protein-coupled receptor 83 [Source:HGNC Symbol;Acc:HGNC:4523] | 17610 | 0.000 | -0.4130 | No |
| 24 | TECTB | tectorin beta [Source:HGNC Symbol;Acc:HGNC:11721] | 17880 | 0.000 | -0.4203 | No |
| 25 | SEMA3G | semaphorin 3G [Source:HGNC Symbol;Acc:HGNC:30400] | 17909 | 0.000 | -0.4210 | No |
| 26 | DUTP6 | deoxyuridine triphosphatase pseudogene 6 [Source:HGNC Symbol;Acc:HGNC:39519] | 18473 | 0.000 | -0.4362 | No |
| 27 | STARD13 | StAR related lipid transfer domain containing 13 [Source:HGNC Symbol;Acc:HGNC:19164] | 19931 | 0.000 | -0.4756 | No |
| 28 | SCN7A | sodium voltage-gated channel alpha subunit 7 [Source:HGNC Symbol;Acc:HGNC:10594] | 20395 | 0.000 | -0.4881 | No |
| 29 | CHL1-AS2 | CHL1 antisense RNA 2 [Source:HGNC Symbol;Acc:HGNC:40147] | 20806 | 0.000 | -0.4991 | No |
| 30 | BMP8B | bone morphogenetic protein 8b [Source:HGNC Symbol;Acc:HGNC:1075] | 20943 | 0.000 | -0.5028 | No |
| 31 | GJC3 | gap junction protein gamma 3 [Source:HGNC Symbol;Acc:HGNC:17495] | 21026 | 0.000 | -0.5050 | No |
| 32 | EDNRB | endothelin receptor type B [Source:HGNC Symbol;Acc:HGNC:3180] | 22020 | 0.000 | -0.5318 | No |
| 33 | NANOGP1 | Nanog homeobox pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:23099] | 23351 | 0.000 | -0.5678 | No |
| 34 | FOXD3 | forkhead box D3 [Source:HGNC Symbol;Acc:HGNC:3804] | 23845 | 0.000 | -0.5811 | No |
| 35 | LINC00466 | long intergenic non-protein coding RNA 466 [Source:HGNC Symbol;Acc:HGNC:27294] | 24857 | 0.000 | -0.6084 | No |
| 36 | KIAA1755 | KIAA1755 [Source:HGNC Symbol;Acc:HGNC:29372] | 26090 | 0.000 | -0.6416 | No |
| 37 | OLFML2A | olfactomedin like 2A [Source:HGNC Symbol;Acc:HGNC:27270] | 26663 | 0.000 | -0.6571 | No |
| 38 | ITIH6 | inter-alpha-trypsin inhibitor heavy chain family member 6 [Source:HGNC Symbol;Acc:HGNC:28907] | 27204 | 0.000 | -0.6716 | No |
| 39 | GAS2L3 | growth arrest specific 2 like 3 [Source:HGNC Symbol;Acc:HGNC:27475] | 27856 | -0.010 | -0.6886 | No |
| 40 | ABCA8 | ATP binding cassette subfamily A member 8 [Source:HGNC Symbol;Acc:HGNC:38] | 28177 | -0.060 | -0.6937 | No |
| 41 | RHBDF1 | rhomboid 5 homolog 1 [Source:HGNC Symbol;Acc:HGNC:20561] | 28937 | -0.163 | -0.7044 | No |
| 42 | C12orf76 | chromosome 12 open reading frame 76 [Source:HGNC Symbol;Acc:HGNC:33790] | 31276 | -0.507 | -0.7371 | Yes |
| 43 | MPZ | myelin protein zero [Source:HGNC Symbol;Acc:HGNC:7225] | 32551 | -0.758 | -0.7261 | Yes |
| 44 | TMEM176A | transmembrane protein 176A [Source:HGNC Symbol;Acc:HGNC:24930] | 32868 | -0.838 | -0.6844 | Yes |
| 45 | PLP1 | proteolipid protein 1 [Source:HGNC Symbol;Acc:HGNC:9086] | 33144 | -0.912 | -0.6372 | Yes |
| 46 | C1orf198 | chromosome 1 open reading frame 198 [Source:HGNC Symbol;Acc:HGNC:25900] | 33172 | -0.919 | -0.5828 | Yes |
| 47 | CADM4 | cell adhesion molecule 4 [Source:HGNC Symbol;Acc:HGNC:30825] | 33642 | -1.061 | -0.5318 | Yes |
| 48 | CNP | 2',3'-cyclic nucleotide 3' phosphodiesterase [Source:HGNC Symbol;Acc:HGNC:2158] | 35073 | -1.611 | -0.4738 | Yes |
| 49 | PRNP | prion protein [Source:HGNC Symbol;Acc:HGNC:9449] | 35495 | -1.834 | -0.3753 | Yes |
| 50 | PMP22 | peripheral myelin protein 22 [Source:HGNC Symbol;Acc:HGNC:9118] | 36760 | -3.182 | -0.2186 | Yes |
| 51 | EXTL1 | exostosin like glycosyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:3515] | 36956 | -3.794 | 0.0036 | Yes |