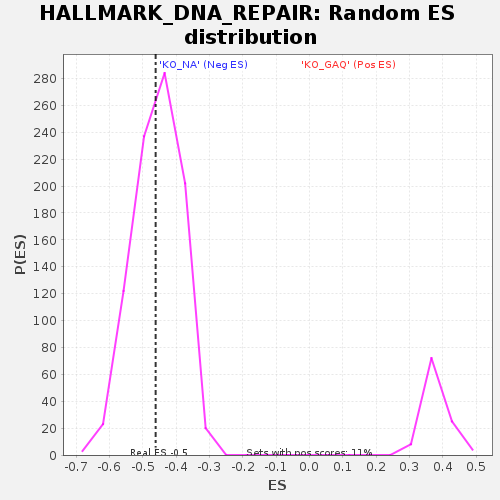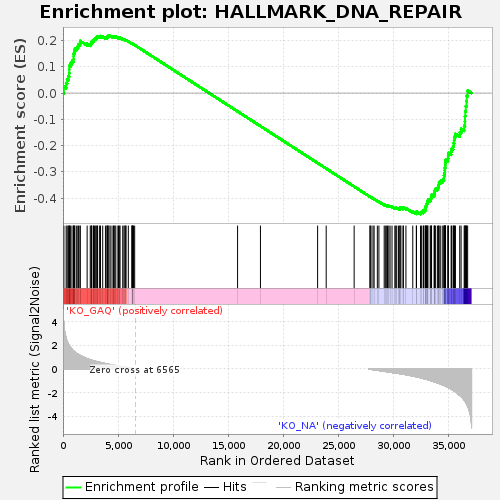
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.4611761 |
| Normalized Enrichment Score (NES) | -1.0092498 |
| Nominal p-value | 0.45679012 |
| FDR q-value | 0.6998329 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PDE4B | phosphodiesterase 4B [Source:HGNC Symbol;Acc:HGNC:8781] | 81 | 3.660 | 0.0241 | No |
| 2 | COX17 | cytochrome c oxidase copper chaperone COX17 [Source:HGNC Symbol;Acc:HGNC:2264] | 286 | 2.608 | 0.0373 | No |
| 3 | PNP | purine nucleoside phosphorylase [Source:HGNC Symbol;Acc:HGNC:7892] | 377 | 2.304 | 0.0514 | No |
| 4 | HCLS1 | hematopoietic cell-specific Lyn substrate 1 [Source:HGNC Symbol;Acc:HGNC:4844] | 482 | 2.111 | 0.0638 | No |
| 5 | POLE4 | DNA polymerase epsilon 4, accessory subunit [Source:HGNC Symbol;Acc:HGNC:18755] | 560 | 1.979 | 0.0759 | No |
| 6 | ERCC1 | ERCC excision repair 1, endonuclease non-catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3433] | 570 | 1.964 | 0.0898 | No |
| 7 | NME1 | NME/NM23 nucleoside diphosphate kinase 1 [Source:HGNC Symbol;Acc:HGNC:7849] | 573 | 1.961 | 0.1038 | No |
| 8 | GTF2H1 | general transcription factor IIH subunit 1 [Source:HGNC Symbol;Acc:HGNC:4655] | 692 | 1.807 | 0.1136 | No |
| 9 | CMPK2 | cytidine/uridine monophosphate kinase 2 [Source:HGNC Symbol;Acc:HGNC:27015] | 818 | 1.639 | 0.1220 | No |
| 10 | APRT | adenine phosphoribosyltransferase [Source:HGNC Symbol;Acc:HGNC:626] | 961 | 1.507 | 0.1290 | No |
| 11 | GMPR2 | guanosine monophosphate reductase 2 [Source:HGNC Symbol;Acc:HGNC:4377] | 963 | 1.506 | 0.1398 | No |
| 12 | GTF2H5 | general transcription factor IIH subunit 5 [Source:HGNC Symbol;Acc:HGNC:21157] | 965 | 1.503 | 0.1505 | No |
| 13 | ELL | elongation factor for RNA polymerase II [Source:HGNC Symbol;Acc:HGNC:23114] | 1036 | 1.444 | 0.1590 | No |
| 14 | ADRM1 | adhesion regulating molecule 1 [Source:HGNC Symbol;Acc:HGNC:15759] | 1078 | 1.411 | 0.1680 | No |
| 15 | ELOA | elongin A [Source:HGNC Symbol;Acc:HGNC:11620] | 1241 | 1.305 | 0.1730 | No |
| 16 | POLR2K | RNA polymerase II subunit K [Source:HGNC Symbol;Acc:HGNC:9198] | 1351 | 1.241 | 0.1790 | No |
| 17 | POLR2J | RNA polymerase II subunit J [Source:HGNC Symbol;Acc:HGNC:9197] | 1415 | 1.196 | 0.1859 | No |
| 18 | DAD1 | defender against cell death 1 [Source:HGNC Symbol;Acc:HGNC:2664] | 1551 | 1.135 | 0.1904 | No |
| 19 | TAF12 | TATA-box binding protein associated factor 12 [Source:HGNC Symbol;Acc:HGNC:11545] | 1558 | 1.133 | 0.1984 | No |
| 20 | ERCC8 | ERCC excision repair 8, CSA ubiquitin ligase complex subunit [Source:HGNC Symbol;Acc:HGNC:3439] | 2191 | 0.866 | 0.1875 | No |
| 21 | ITPA | inosine triphosphatase [Source:HGNC Symbol;Acc:HGNC:6176] | 2472 | 0.774 | 0.1855 | No |
| 22 | TAF13 | TATA-box binding protein associated factor 13 [Source:HGNC Symbol;Acc:HGNC:11546] | 2555 | 0.745 | 0.1886 | No |
| 23 | DGCR8 | DGCR8 microprocessor complex subunit [Source:HGNC Symbol;Acc:HGNC:2847] | 2589 | 0.736 | 0.1930 | No |
| 24 | POLR2C | RNA polymerase II subunit C [Source:HGNC Symbol;Acc:HGNC:9189] | 2614 | 0.728 | 0.1976 | No |
| 25 | SRSF6 | serine and arginine rich splicing factor 6 [Source:HGNC Symbol;Acc:HGNC:10788] | 2760 | 0.692 | 0.1986 | No |
| 26 | GTF2F1 | general transcription factor IIF subunit 1 [Source:HGNC Symbol;Acc:HGNC:4652] | 2780 | 0.687 | 0.2030 | No |
| 27 | SNAPC5 | small nuclear RNA activating complex polypeptide 5 [Source:HGNC Symbol;Acc:HGNC:15484] | 2854 | 0.665 | 0.2058 | No |
| 28 | TAF1C | TATA-box binding protein associated factor, RNA polymerase I subunit C [Source:HGNC Symbol;Acc:HGNC:11534] | 2975 | 0.636 | 0.2072 | No |
| 29 | SNAPC4 | small nuclear RNA activating complex polypeptide 4 [Source:HGNC Symbol;Acc:HGNC:11137] | 3009 | 0.628 | 0.2108 | No |
| 30 | GTF2B | general transcription factor IIB [Source:HGNC Symbol;Acc:HGNC:4648] | 3095 | 0.609 | 0.2129 | No |
| 31 | SEC61A1 | SEC61 translocon subunit alpha 1 [Source:HGNC Symbol;Acc:HGNC:18276] | 3130 | 0.601 | 0.2163 | No |
| 32 | NT5C3A | 5'-nucleotidase, cytosolic IIIA [Source:HGNC Symbol;Acc:HGNC:17820] | 3302 | 0.561 | 0.2157 | No |
| 33 | RRM2B | ribonucleotide reductase regulatory TP53 inducible subunit M2B [Source:HGNC Symbol;Acc:HGNC:17296] | 3404 | 0.535 | 0.2168 | No |
| 34 | POLR1D | RNA polymerase I and III subunit D [Source:HGNC Symbol;Acc:HGNC:20422] | 3604 | 0.492 | 0.2149 | No |
| 35 | ERCC3 | ERCC excision repair 3, TFIIH core complex helicase subunit [Source:HGNC Symbol;Acc:HGNC:3435] | 3852 | 0.444 | 0.2114 | No |
| 36 | SDCBP | syndecan binding protein [Source:HGNC Symbol;Acc:HGNC:10662] | 3994 | 0.416 | 0.2106 | No |
| 37 | TARBP2 | TARBP2 subunit of RISC loading complex [Source:HGNC Symbol;Acc:HGNC:11569] | 4004 | 0.414 | 0.2133 | No |
| 38 | RNMT | RNA guanine-7 methyltransferase [Source:HGNC Symbol;Acc:HGNC:10075] | 4059 | 0.406 | 0.2148 | No |
| 39 | NCBP2 | nuclear cap binding protein subunit 2 [Source:HGNC Symbol;Acc:HGNC:7659] | 4080 | 0.403 | 0.2171 | No |
| 40 | ERCC2 | ERCC excision repair 2, TFIIH core complex helicase subunit [Source:HGNC Symbol;Acc:HGNC:3434] | 4166 | 0.389 | 0.2176 | No |
| 41 | SMAD5 | SMAD family member 5 [Source:HGNC Symbol;Acc:HGNC:6771] | 4282 | 0.365 | 0.2171 | No |
| 42 | EIF1B | eukaryotic translation initiation factor 1B [Source:HGNC Symbol;Acc:HGNC:30792] | 4451 | 0.335 | 0.2150 | No |
| 43 | POLR2D | RNA polymerase II subunit D [Source:HGNC Symbol;Acc:HGNC:9191] | 4591 | 0.312 | 0.2135 | No |
| 44 | POLR2F | RNA polymerase II subunit F [Source:HGNC Symbol;Acc:HGNC:9193] | 4621 | 0.309 | 0.2149 | No |
| 45 | TSG101 | tumor susceptibility 101 [Source:HGNC Symbol;Acc:HGNC:15971] | 4701 | 0.297 | 0.2149 | No |
| 46 | EDF1 | endothelial differentiation related factor 1 [Source:HGNC Symbol;Acc:HGNC:3164] | 4793 | 0.280 | 0.2144 | No |
| 47 | POLB | DNA polymerase beta [Source:HGNC Symbol;Acc:HGNC:9174] | 4985 | 0.247 | 0.2110 | No |
| 48 | GTF2H3 | general transcription factor IIH subunit 3 [Source:HGNC Symbol;Acc:HGNC:4657] | 5037 | 0.235 | 0.2114 | No |
| 49 | TMED2 | transmembrane p24 trafficking protein 2 [Source:HGNC Symbol;Acc:HGNC:16996] | 5153 | 0.214 | 0.2098 | No |
| 50 | CANT1 | calcium activated nucleotidase 1 [Source:HGNC Symbol;Acc:HGNC:19721] | 5165 | 0.212 | 0.2110 | No |
| 51 | SF3A3 | splicing factor 3a subunit 3 [Source:HGNC Symbol;Acc:HGNC:10767] | 5422 | 0.173 | 0.2053 | No |
| 52 | BRF2 | BRF2 RNA polymerase III transcription initiation factor subunit [Source:HGNC Symbol;Acc:HGNC:17298] | 5535 | 0.156 | 0.2034 | No |
| 53 | CSTF3 | cleavage stimulation factor subunit 3 [Source:HGNC Symbol;Acc:HGNC:2485] | 5623 | 0.143 | 0.2021 | No |
| 54 | RALA | RAS like proto-oncogene A [Source:HGNC Symbol;Acc:HGNC:9839] | 5702 | 0.136 | 0.2009 | No |
| 55 | VPS37B | VPS37B subunit of ESCRT-I [Source:HGNC Symbol;Acc:HGNC:25754] | 5710 | 0.135 | 0.2017 | No |
| 56 | POLR2G | RNA polymerase II subunit G [Source:HGNC Symbol;Acc:HGNC:9194] | 5930 | 0.101 | 0.1965 | No |
| 57 | POLD4 | DNA polymerase delta 4, accessory subunit [Source:HGNC Symbol;Acc:HGNC:14106] | 6264 | 0.047 | 0.1878 | No |
| 58 | RBX1 | ring-box 1 [Source:HGNC Symbol;Acc:HGNC:9928] | 6274 | 0.045 | 0.1879 | No |
| 59 | CETN2 | centrin 2 [Source:HGNC Symbol;Acc:HGNC:1867] | 6291 | 0.043 | 0.1878 | No |
| 60 | UMPS | uridine monophosphate synthetase [Source:HGNC Symbol;Acc:HGNC:12563] | 6300 | 0.042 | 0.1879 | No |
| 61 | CLP1 | cleavage factor polyribonucleotide kinase subunit 1 [Source:HGNC Symbol;Acc:HGNC:16999] | 6346 | 0.033 | 0.1869 | No |
| 62 | PDE6G | phosphodiesterase 6G [Source:HGNC Symbol;Acc:HGNC:8789] | 6402 | 0.026 | 0.1856 | No |
| 63 | POM121 | POM121 transmembrane nucleoporin [Source:HGNC Symbol;Acc:HGNC:19702] | 6506 | 0.009 | 0.1829 | No |
| 64 | BCAM | basal cell adhesion molecule (Lutheran blood group) [Source:HGNC Symbol;Acc:HGNC:6722] | 15848 | 0.000 | -0.0700 | No |
| 65 | BOLA2 | bolA family member 2 [Source:HGNC Symbol;Acc:HGNC:29488] | 17924 | 0.000 | -0.1262 | No |
| 66 | CCNO | cyclin O [Source:HGNC Symbol;Acc:HGNC:18576] | 23114 | 0.000 | -0.2666 | No |
| 67 | NPR2 | natriuretic peptide receptor 2 [Source:HGNC Symbol;Acc:HGNC:7944] | 23892 | 0.000 | -0.2876 | No |
| 68 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26430 | 0.000 | -0.3563 | No |
| 69 | SUPT4H1 | SPT4 homolog, DSIF elongation factor subunit [Source:HGNC Symbol;Acc:HGNC:11467] | 27868 | -0.012 | -0.3951 | No |
| 70 | SUPT5H | SPT5 homolog, DSIF elongation factor subunit [Source:HGNC Symbol;Acc:HGNC:11469] | 27881 | -0.014 | -0.3954 | No |
| 71 | CDA | cytidine deaminase [Source:HGNC Symbol;Acc:HGNC:1712] | 27961 | -0.029 | -0.3973 | No |
| 72 | DDB1 | damage specific DNA binding protein 1 [Source:HGNC Symbol;Acc:HGNC:2717] | 28140 | -0.054 | -0.4017 | No |
| 73 | SAC3D1 | SAC3 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:30179] | 28242 | -0.069 | -0.4040 | No |
| 74 | POLA2 | DNA polymerase alpha 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:30073] | 28535 | -0.107 | -0.4111 | No |
| 75 | NUDT9 | nudix hydrolase 9 [Source:HGNC Symbol;Acc:HGNC:8056] | 28677 | -0.129 | -0.4140 | No |
| 76 | MRPL40 | mitochondrial ribosomal protein L40 [Source:HGNC Symbol;Acc:HGNC:14491] | 29158 | -0.193 | -0.4256 | No |
| 77 | TAF9 | TATA-box binding protein associated factor 9 [Source:HGNC Symbol;Acc:HGNC:11542] | 29260 | -0.211 | -0.4268 | No |
| 78 | NELFB | negative elongation factor complex member B [Source:HGNC Symbol;Acc:HGNC:24324] | 29384 | -0.229 | -0.4285 | No |
| 79 | TAF10 | TATA-box binding protein associated factor 10 [Source:HGNC Symbol;Acc:HGNC:11543] | 29460 | -0.239 | -0.4288 | No |
| 80 | STX3 | syntaxin 3 [Source:HGNC Symbol;Acc:HGNC:11438] | 29576 | -0.253 | -0.4301 | No |
| 81 | GTF2A2 | general transcription factor IIA subunit 2 [Source:HGNC Symbol;Acc:HGNC:4647] | 29692 | -0.271 | -0.4313 | No |
| 82 | UPF3B | UPF3B regulator of nonsense mediated mRNA decay [Source:HGNC Symbol;Acc:HGNC:20439] | 29868 | -0.299 | -0.4339 | No |
| 83 | NFX1 | nuclear transcription factor, X-box binding 1 [Source:HGNC Symbol;Acc:HGNC:7803] | 30124 | -0.332 | -0.4384 | No |
| 84 | MPC2 | mitochondrial pyruvate carrier 2 [Source:HGNC Symbol;Acc:HGNC:24515] | 30206 | -0.342 | -0.4381 | No |
| 85 | BCAP31 | B cell receptor associated protein 31 [Source:HGNC Symbol;Acc:HGNC:16695] | 30294 | -0.354 | -0.4379 | No |
| 86 | FEN1 | flap structure-specific endonuclease 1 [Source:HGNC Symbol;Acc:HGNC:3650] | 30452 | -0.375 | -0.4395 | No |
| 87 | RAE1 | ribonucleic acid export 1 [Source:HGNC Symbol;Acc:HGNC:9828] | 30579 | -0.397 | -0.4400 | No |
| 88 | POLR2H | RNA polymerase II subunit H [Source:HGNC Symbol;Acc:HGNC:9195] | 30587 | -0.399 | -0.4374 | No |
| 89 | POLR3C | RNA polymerase III subunit C [Source:HGNC Symbol;Acc:HGNC:30076] | 30674 | -0.413 | -0.4367 | No |
| 90 | ERCC5 | ERCC excision repair 5, endonuclease [Source:HGNC Symbol;Acc:HGNC:3437] | 30866 | -0.444 | -0.4387 | No |
| 91 | SSRP1 | structure specific recognition protein 1 [Source:HGNC Symbol;Acc:HGNC:11327] | 30899 | -0.449 | -0.4363 | No |
| 92 | POLR2I | RNA polymerase II subunit I [Source:HGNC Symbol;Acc:HGNC:9196] | 31131 | -0.482 | -0.4391 | No |
| 93 | AK1 | adenylate kinase 1 [Source:HGNC Symbol;Acc:HGNC:361] | 31752 | -0.588 | -0.4517 | No |
| 94 | NELFE | negative elongation factor complex member E [Source:HGNC Symbol;Acc:HGNC:13974] | 32074 | -0.657 | -0.4557 | No |
| 95 | USP11 | ubiquitin specific peptidase 11 [Source:HGNC Symbol;Acc:HGNC:12609] | 32091 | -0.660 | -0.4513 | No |
| 96 | NUDT21 | nudix hydrolase 21 [Source:HGNC Symbol;Acc:HGNC:13870] | 32455 | -0.734 | -0.4559 | Yes |
| 97 | IMPDH2 | inosine monophosphate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:6053] | 32534 | -0.752 | -0.4526 | Yes |
| 98 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32700 | -0.796 | -0.4514 | Yes |
| 99 | POLR2E | RNA polymerase II subunit E [Source:HGNC Symbol;Acc:HGNC:9192] | 32720 | -0.802 | -0.4461 | Yes |
| 100 | POLR1C | RNA polymerase I and III subunit C [Source:HGNC Symbol;Acc:HGNC:20194] | 32874 | -0.840 | -0.4442 | Yes |
| 101 | AAAS | aladin WD repeat nucleoporin [Source:HGNC Symbol;Acc:HGNC:13666] | 32892 | -0.845 | -0.4386 | Yes |
| 102 | NT5C | 5', 3'-nucleotidase, cytosolic [Source:HGNC Symbol;Acc:HGNC:17144] | 32910 | -0.849 | -0.4330 | Yes |
| 103 | ERCC4 | ERCC excision repair 4, endonuclease catalytic subunit [Source:HGNC Symbol;Acc:HGNC:3436] | 32964 | -0.865 | -0.4282 | Yes |
| 104 | HPRT1 | hypoxanthine phosphoribosyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:5157] | 33002 | -0.873 | -0.4229 | Yes |
| 105 | ALYREF | Aly/REF export factor [Source:HGNC Symbol;Acc:HGNC:19071] | 33064 | -0.887 | -0.4182 | Yes |
| 106 | ARL6IP1 | ADP ribosylation factor like GTPase 6 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:697] | 33076 | -0.891 | -0.4121 | Yes |
| 107 | ADCY6 | adenylate cyclase 6 [Source:HGNC Symbol;Acc:HGNC:237] | 33132 | -0.907 | -0.4071 | Yes |
| 108 | NELFCD | negative elongation factor complex member C/D [Source:HGNC Symbol;Acc:HGNC:15934] | 33335 | -0.967 | -0.4056 | Yes |
| 109 | DCTN4 | dynactin subunit 4 [Source:HGNC Symbol;Acc:HGNC:15518] | 33401 | -0.987 | -0.4003 | Yes |
| 110 | POLR2A | RNA polymerase II subunit A [Source:HGNC Symbol;Acc:HGNC:9187] | 33425 | -0.996 | -0.3937 | Yes |
| 111 | RAD52 | RAD52 homolog, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:9824] | 33457 | -1.006 | -0.3873 | Yes |
| 112 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33690 | -1.077 | -0.3859 | Yes |
| 113 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.3789 | Yes |
| 114 | VPS28 | VPS28 subunit of ESCRT-I [Source:HGNC Symbol;Acc:HGNC:18178] | 33741 | -1.090 | -0.3716 | Yes |
| 115 | TK2 | thymidine kinase 2 [Source:HGNC Symbol;Acc:HGNC:11831] | 33802 | -1.112 | -0.3652 | Yes |
| 116 | VPS37D | VPS37D subunit of ESCRT-I [Source:HGNC Symbol;Acc:HGNC:18287] | 33984 | -1.180 | -0.3616 | Yes |
| 117 | TAF6 | TATA-box binding protein associated factor 6 [Source:HGNC Symbol;Acc:HGNC:11540] | 34063 | -1.203 | -0.3551 | Yes |
| 118 | TP53 | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 34112 | -1.214 | -0.3477 | Yes |
| 119 | MPG | N-methylpurine DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7211] | 34143 | -1.223 | -0.3397 | Yes |
| 120 | NME3 | NME/NM23 nucleoside diphosphate kinase 3 [Source:HGNC Symbol;Acc:HGNC:7851] | 34292 | -1.278 | -0.3345 | Yes |
| 121 | POLH | DNA polymerase eta [Source:HGNC Symbol;Acc:HGNC:9181] | 34476 | -1.343 | -0.3298 | Yes |
| 122 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 34584 | -1.386 | -0.3228 | Yes |
| 123 | DGUOK | deoxyguanosine kinase [Source:HGNC Symbol;Acc:HGNC:2858] | 34589 | -1.387 | -0.3129 | Yes |
| 124 | GSDME | gasdermin E [Source:HGNC Symbol;Acc:HGNC:2810] | 34644 | -1.408 | -0.3043 | Yes |
| 125 | POLL | DNA polymerase lambda [Source:HGNC Symbol;Acc:HGNC:9184] | 34660 | -1.414 | -0.2945 | Yes |
| 126 | SURF1 | SURF1 cytochrome c oxidase assembly factor [Source:HGNC Symbol;Acc:HGNC:11474] | 34667 | -1.417 | -0.2845 | Yes |
| 127 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34705 | -1.428 | -0.2753 | Yes |
| 128 | ZNF707 | zinc finger protein 707 [Source:HGNC Symbol;Acc:HGNC:27815] | 34709 | -1.429 | -0.2651 | Yes |
| 129 | XPC | XPC complex subunit, DNA damage recognition and repair factor [Source:HGNC Symbol;Acc:HGNC:12816] | 34727 | -1.435 | -0.2552 | Yes |
| 130 | GUK1 | guanylate kinase 1 [Source:HGNC Symbol;Acc:HGNC:4693] | 34940 | -1.536 | -0.2499 | Yes |
| 131 | GTF3C5 | general transcription factor IIIC subunit 5 [Source:HGNC Symbol;Acc:HGNC:4668] | 34965 | -1.551 | -0.2394 | Yes |
| 132 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 34976 | -1.554 | -0.2285 | Yes |
| 133 | RPA2 | replication protein A2 [Source:HGNC Symbol;Acc:HGNC:10290] | 35234 | -1.694 | -0.2233 | Yes |
| 134 | AGO4 | argonaute RISC component 4 [Source:HGNC Symbol;Acc:HGNC:18424] | 35279 | -1.719 | -0.2122 | Yes |
| 135 | RAD51 | RAD51 recombinase [Source:HGNC Symbol;Acc:HGNC:9817] | 35417 | -1.787 | -0.2030 | Yes |
| 136 | POLA1 | DNA polymerase alpha 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9173] | 35456 | -1.812 | -0.1910 | Yes |
| 137 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35508 | -1.842 | -0.1792 | Yes |
| 138 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 35534 | -1.859 | -0.1665 | Yes |
| 139 | ADA | adenosine deaminase [Source:HGNC Symbol;Acc:HGNC:186] | 35626 | -1.922 | -0.1552 | Yes |
| 140 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 36015 | -2.212 | -0.1498 | Yes |
| 141 | NME4 | NME/NM23 nucleoside diphosphate kinase 4 [Source:HGNC Symbol;Acc:HGNC:7852] | 36138 | -2.314 | -0.1365 | Yes |
| 142 | RFC2 | replication factor C subunit 2 [Source:HGNC Symbol;Acc:HGNC:9970] | 36418 | -2.602 | -0.1253 | Yes |
| 143 | AK3 | adenylate kinase 3 [Source:HGNC Symbol;Acc:HGNC:17376] | 36486 | -2.700 | -0.1077 | Yes |
| 144 | REV3L | REV3 like, DNA directed polymerase zeta catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9968] | 36493 | -2.706 | -0.0885 | Yes |
| 145 | DDB2 | damage specific DNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:2718] | 36524 | -2.760 | -0.0694 | Yes |
| 146 | POLR3GL | RNA polymerase III subunit G like [Source:HGNC Symbol;Acc:HGNC:28466] | 36563 | -2.815 | -0.0502 | Yes |
| 147 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36627 | -2.912 | -0.0310 | Yes |
| 148 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36649 | -2.950 | -0.0104 | Yes |
| 149 | GPX4 | glutathione peroxidase 4 [Source:HGNC Symbol;Acc:HGNC:4556] | 36733 | -3.106 | 0.0097 | Yes |