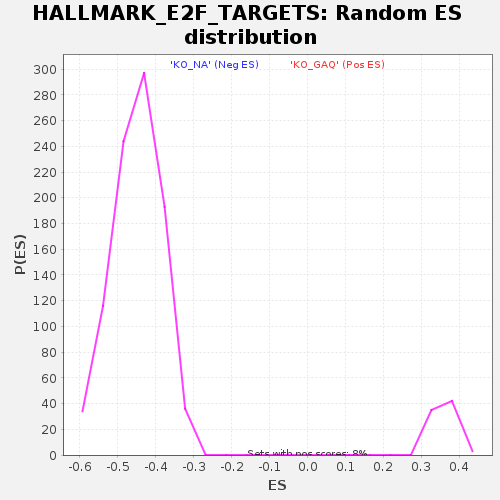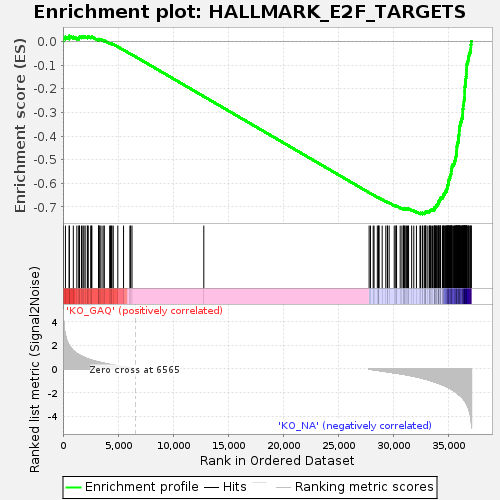
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_E2F_TARGETS |
| Enrichment Score (ES) | -0.7301012 |
| Normalized Enrichment Score (NES) | -1.6237581 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0013035353 |
| FWER p-Value | 0.002 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MYC | MYC proto-oncogene, bHLH transcription factor [Source:HGNC Symbol;Acc:HGNC:7553] | 18 | 4.578 | 0.0151 | No |
| 2 | TFRC | transferrin receptor [Source:HGNC Symbol;Acc:HGNC:11763] | 221 | 2.836 | 0.0193 | No |
| 3 | POLE4 | DNA polymerase epsilon 4, accessory subunit [Source:HGNC Symbol;Acc:HGNC:18755] | 560 | 1.979 | 0.0169 | No |
| 4 | NME1 | NME/NM23 nucleoside diphosphate kinase 1 [Source:HGNC Symbol;Acc:HGNC:7849] | 573 | 1.961 | 0.0233 | No |
| 5 | HUS1 | HUS1 checkpoint clamp component [Source:HGNC Symbol;Acc:HGNC:5309] | 937 | 1.528 | 0.0187 | No |
| 6 | UBE2S | ubiquitin conjugating enzyme E2 S [Source:HGNC Symbol;Acc:HGNC:17895] | 1258 | 1.294 | 0.0144 | No |
| 7 | PA2G4 | proliferation-associated 2G4 [Source:HGNC Symbol;Acc:HGNC:8550] | 1435 | 1.187 | 0.0137 | No |
| 8 | EIF2S1 | eukaryotic translation initiation factor 2 subunit alpha [Source:HGNC Symbol;Acc:HGNC:3265] | 1473 | 1.164 | 0.0167 | No |
| 9 | CKS2 | CDC28 protein kinase regulatory subunit 2 [Source:HGNC Symbol;Acc:HGNC:2000] | 1504 | 1.154 | 0.0198 | No |
| 10 | SYNCRIP | synaptotagmin binding cytoplasmic RNA interacting protein [Source:HGNC Symbol;Acc:HGNC:16918] | 1680 | 1.074 | 0.0187 | No |
| 11 | NOP56 | NOP56 ribonucleoprotein [Source:HGNC Symbol;Acc:HGNC:15911] | 1806 | 1.013 | 0.0188 | No |
| 12 | ORC2 | origin recognition complex subunit 2 [Source:HGNC Symbol;Acc:HGNC:8488] | 1895 | 0.982 | 0.0197 | No |
| 13 | DDX39A | DExD-box helicase 39A [Source:HGNC Symbol;Acc:HGNC:17821] | 2010 | 0.931 | 0.0198 | No |
| 14 | NUP153 | nucleoporin 153 [Source:HGNC Symbol;Acc:HGNC:8062] | 2206 | 0.861 | 0.0175 | No |
| 15 | RANBP1 | RAN binding protein 1 [Source:HGNC Symbol;Acc:HGNC:9847] | 2244 | 0.846 | 0.0194 | No |
| 16 | NOLC1 | nucleolar and coiled-body phosphoprotein 1 [Source:HGNC Symbol;Acc:HGNC:15608] | 2285 | 0.832 | 0.0211 | No |
| 17 | SRSF1 | serine and arginine rich splicing factor 1 [Source:HGNC Symbol;Acc:HGNC:10780] | 2515 | 0.757 | 0.0175 | No |
| 18 | POP7 | POP7 homolog, ribonuclease P/MRP subunit [Source:HGNC Symbol;Acc:HGNC:19949] | 2595 | 0.735 | 0.0179 | No |
| 19 | RAN | RAN, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:9846] | 2617 | 0.727 | 0.0198 | No |
| 20 | CDC25A | cell division cycle 25A [Source:HGNC Symbol;Acc:HGNC:1725] | 3222 | 0.578 | 0.0054 | No |
| 21 | SNRPB | small nuclear ribonucleoprotein polypeptides B and B1 [Source:HGNC Symbol;Acc:HGNC:11153] | 3239 | 0.573 | 0.0069 | No |
| 22 | TRA2B | transformer 2 beta homolog [Source:HGNC Symbol;Acc:HGNC:10781] | 3246 | 0.571 | 0.0087 | No |
| 23 | HMGA1 | high mobility group AT-hook 1 [Source:HGNC Symbol;Acc:HGNC:5010] | 3283 | 0.564 | 0.0096 | No |
| 24 | EED | embryonic ectoderm development [Source:HGNC Symbol;Acc:HGNC:3188] | 3379 | 0.542 | 0.0089 | No |
| 25 | GSPT1 | G1 to S phase transition 1 [Source:HGNC Symbol;Acc:HGNC:4621] | 3543 | 0.504 | 0.0062 | No |
| 26 | CDKN1A | cyclin dependent kinase inhibitor 1A [Source:HGNC Symbol;Acc:HGNC:1784] | 3722 | 0.467 | 0.0030 | No |
| 27 | LYAR | Ly1 antibody reactive [Source:HGNC Symbol;Acc:HGNC:26021] | 3737 | 0.465 | 0.0042 | No |
| 28 | PRPS1 | phosphoribosyl pyrophosphate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:9462] | 4236 | 0.373 | -0.0080 | No |
| 29 | SRSF2 | serine and arginine rich splicing factor 2 [Source:HGNC Symbol;Acc:HGNC:10783] | 4311 | 0.359 | -0.0088 | No |
| 30 | DCTPP1 | dCTP pyrophosphatase 1 [Source:HGNC Symbol;Acc:HGNC:28777] | 4412 | 0.341 | -0.0104 | No |
| 31 | MTHFD2 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase [Source:HGNC Symbol;Acc:HGNC:7434] | 4439 | 0.337 | -0.0099 | No |
| 32 | DONSON | DNA replication fork stabilization factor DONSON [Source:HGNC Symbol;Acc:HGNC:2993] | 4562 | 0.315 | -0.0122 | No |
| 33 | HNRNPD | heterogeneous nuclear ribonucleoprotein D [Source:HGNC Symbol;Acc:HGNC:5036] | 4976 | 0.248 | -0.0225 | No |
| 34 | STAG1 | stromal antigen 1 [Source:HGNC Symbol;Acc:HGNC:11354] | 5496 | 0.163 | -0.0360 | No |
| 35 | NAA38 | N-alpha-acetyltransferase 38, NatC auxiliary subunit [Source:HGNC Symbol;Acc:HGNC:28212] | 6075 | 0.084 | -0.0514 | No |
| 36 | TBRG4 | transforming growth factor beta regulator 4 [Source:HGNC Symbol;Acc:HGNC:17443] | 6140 | 0.072 | -0.0529 | No |
| 37 | AK2 | adenylate kinase 2 [Source:HGNC Symbol;Acc:HGNC:362] | 6263 | 0.047 | -0.0560 | No |
| 38 | CDKN2A | cyclin dependent kinase inhibitor 2A [Source:HGNC Symbol;Acc:HGNC:1787] | 12782 | 0.000 | -0.2327 | No |
| 39 | PHF5A | PHD finger protein 5A [Source:HGNC Symbol;Acc:HGNC:18000] | 27790 | -0.002 | -0.6395 | No |
| 40 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 27912 | -0.021 | -0.6427 | No |
| 41 | JPT1 | Jupiter microtubule associated homolog 1 [Source:HGNC Symbol;Acc:HGNC:14569] | 28159 | -0.057 | -0.6492 | No |
| 42 | SHMT1 | serine hydroxymethyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:10850] | 28235 | -0.069 | -0.6510 | No |
| 43 | POLA2 | DNA polymerase alpha 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:30073] | 28535 | -0.107 | -0.6587 | No |
| 44 | PTTG1 | PTTG1 regulator of sister chromatid separation, securin [Source:HGNC Symbol;Acc:HGNC:9690] | 28646 | -0.123 | -0.6613 | No |
| 45 | LUC7L3 | LUC7 like 3 pre-mRNA splicing factor [Source:HGNC Symbol;Acc:HGNC:24309] | 28669 | -0.128 | -0.6614 | No |
| 46 | NBN | nibrin [Source:HGNC Symbol;Acc:HGNC:7652] | 28683 | -0.130 | -0.6613 | No |
| 47 | ANP32E | acidic nuclear phosphoprotein 32 family member E [Source:HGNC Symbol;Acc:HGNC:16673] | 28974 | -0.167 | -0.6686 | No |
| 48 | IPO7 | importin 7 [Source:HGNC Symbol;Acc:HGNC:9852] | 29304 | -0.216 | -0.6768 | No |
| 49 | PNN | pinin, desmosome associated protein [Source:HGNC Symbol;Acc:HGNC:9162] | 29447 | -0.238 | -0.6799 | No |
| 50 | NAP1L1 | nucleosome assembly protein 1 like 1 [Source:HGNC Symbol;Acc:HGNC:7637] | 29462 | -0.239 | -0.6794 | No |
| 51 | RBBP7 | RB binding protein 7, chromatin remodeling factor [Source:HGNC Symbol;Acc:HGNC:9890] | 29608 | -0.259 | -0.6825 | No |
| 52 | SLBP | stem-loop binding protein [Source:HGNC Symbol;Acc:HGNC:10904] | 30070 | -0.326 | -0.6939 | No |
| 53 | RAD1 | RAD1 checkpoint DNA exonuclease [Source:HGNC Symbol;Acc:HGNC:9806] | 30218 | -0.343 | -0.6967 | No |
| 54 | PPP1R8 | protein phosphatase 1 regulatory subunit 8 [Source:HGNC Symbol;Acc:HGNC:9296] | 30248 | -0.347 | -0.6963 | No |
| 55 | NUP205 | nucleoporin 205 [Source:HGNC Symbol;Acc:HGNC:18658] | 30281 | -0.352 | -0.6959 | No |
| 56 | CTCF | CCCTC-binding factor [Source:HGNC Symbol;Acc:HGNC:13723] | 30609 | -0.404 | -0.7034 | No |
| 57 | ILF3 | interleukin enhancer binding factor 3 [Source:HGNC Symbol;Acc:HGNC:6038] | 30768 | -0.428 | -0.7062 | No |
| 58 | SSRP1 | structure specific recognition protein 1 [Source:HGNC Symbol;Acc:HGNC:11327] | 30899 | -0.449 | -0.7082 | No |
| 59 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 30914 | -0.450 | -0.7071 | No |
| 60 | CNOT9 | CCR4-NOT transcription complex subunit 9 [Source:HGNC Symbol;Acc:HGNC:10445] | 30963 | -0.457 | -0.7068 | No |
| 61 | ASF1A | anti-silencing function 1A histone chaperone [Source:HGNC Symbol;Acc:HGNC:20995] | 30987 | -0.460 | -0.7059 | No |
| 62 | DCK | deoxycytidine kinase [Source:HGNC Symbol;Acc:HGNC:2704] | 31121 | -0.480 | -0.7078 | No |
| 63 | MLH1 | mutL homolog 1 [Source:HGNC Symbol;Acc:HGNC:7127] | 31170 | -0.489 | -0.7075 | No |
| 64 | PRDX4 | peroxiredoxin 4 [Source:HGNC Symbol;Acc:HGNC:17169] | 31269 | -0.505 | -0.7084 | No |
| 65 | CTPS1 | CTP synthase 1 [Source:HGNC Symbol;Acc:HGNC:2519] | 31341 | -0.517 | -0.7086 | No |
| 66 | DCLRE1B | DNA cross-link repair 1B [Source:HGNC Symbol;Acc:HGNC:17641] | 31371 | -0.521 | -0.7076 | No |
| 67 | ING3 | inhibitor of growth family member 3 [Source:HGNC Symbol;Acc:HGNC:14587] | 31651 | -0.569 | -0.7132 | No |
| 68 | ZW10 | zw10 kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13194] | 31845 | -0.605 | -0.7164 | No |
| 69 | KPNA2 | karyopherin subunit alpha 2 [Source:HGNC Symbol;Acc:HGNC:6395] | 32085 | -0.659 | -0.7206 | No |
| 70 | TIPIN | TIMELESS interacting protein [Source:HGNC Symbol;Acc:HGNC:30750] | 32406 | -0.721 | -0.7268 | No |
| 71 | NUDT21 | nudix hydrolase 21 [Source:HGNC Symbol;Acc:HGNC:13870] | 32455 | -0.734 | -0.7256 | No |
| 72 | PAN2 | poly(A) specific ribonuclease subunit PAN2 [Source:HGNC Symbol;Acc:HGNC:20074] | 32622 | -0.776 | -0.7275 | Yes |
| 73 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 32625 | -0.776 | -0.7249 | Yes |
| 74 | CKS1B | CDC28 protein kinase regulatory subunit 1B [Source:HGNC Symbol;Acc:HGNC:19083] | 32783 | -0.819 | -0.7263 | Yes |
| 75 | LMNB1 | lamin B1 [Source:HGNC Symbol;Acc:HGNC:6637] | 32854 | -0.836 | -0.7254 | Yes |
| 76 | PRKDC | protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413] | 32880 | -0.841 | -0.7232 | Yes |
| 77 | NUP107 | nucleoporin 107 [Source:HGNC Symbol;Acc:HGNC:29914] | 32909 | -0.849 | -0.7210 | Yes |
| 78 | RAD50 | RAD50 double strand break repair protein [Source:HGNC Symbol;Acc:HGNC:9816] | 32927 | -0.853 | -0.7186 | Yes |
| 79 | POLD2 | DNA polymerase delta 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9176] | 33057 | -0.887 | -0.7191 | Yes |
| 80 | PSMC3IP | PSMC3 interacting protein [Source:HGNC Symbol;Acc:HGNC:17928] | 33230 | -0.941 | -0.7205 | Yes |
| 81 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 33267 | -0.949 | -0.7182 | Yes |
| 82 | XRCC6 | X-ray repair cross complementing 6 [Source:HGNC Symbol;Acc:HGNC:4055] | 33316 | -0.961 | -0.7163 | Yes |
| 83 | XPO1 | exportin 1 [Source:HGNC Symbol;Acc:HGNC:12825] | 33352 | -0.972 | -0.7139 | Yes |
| 84 | WDR90 | WD repeat domain 90 [Source:HGNC Symbol;Acc:HGNC:26960] | 33427 | -0.997 | -0.7125 | Yes |
| 85 | DEPDC1 | DEP domain containing 1 [Source:HGNC Symbol;Acc:HGNC:22949] | 33541 | -1.033 | -0.7120 | Yes |
| 86 | RFC3 | replication factor C subunit 3 [Source:HGNC Symbol;Acc:HGNC:9971] | 33690 | -1.077 | -0.7124 | Yes |
| 87 | PLK4 | polo like kinase 4 [Source:HGNC Symbol;Acc:HGNC:11397] | 33697 | -1.078 | -0.7089 | Yes |
| 88 | SPC24 | SPC24 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:26913] | 33701 | -1.079 | -0.7053 | Yes |
| 89 | PCNA | proliferating cell nuclear antigen [Source:HGNC Symbol;Acc:HGNC:8729] | 33721 | -1.084 | -0.7021 | Yes |
| 90 | SMC6 | structural maintenance of chromosomes 6 [Source:HGNC Symbol;Acc:HGNC:20466] | 33822 | -1.119 | -0.7010 | Yes |
| 91 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 33838 | -1.126 | -0.6975 | Yes |
| 92 | TRIP13 | thyroid hormone receptor interactor 13 [Source:HGNC Symbol;Acc:HGNC:12307] | 33877 | -1.138 | -0.6947 | Yes |
| 93 | MAD2L1 | mitotic arrest deficient 2 like 1 [Source:HGNC Symbol;Acc:HGNC:6763] | 33947 | -1.167 | -0.6926 | Yes |
| 94 | TACC3 | transforming acidic coiled-coil containing protein 3 [Source:HGNC Symbol;Acc:HGNC:11524] | 33954 | -1.169 | -0.6887 | Yes |
| 95 | BRMS1L | BRMS1 like transcriptional repressor [Source:HGNC Symbol;Acc:HGNC:20512] | 34054 | -1.200 | -0.6873 | Yes |
| 96 | GINS4 | GINS complex subunit 4 [Source:HGNC Symbol;Acc:HGNC:28226] | 34077 | -1.206 | -0.6838 | Yes |
| 97 | SUV39H1 | suppressor of variegation 3-9 homolog 1 [Source:HGNC Symbol;Acc:HGNC:11479] | 34089 | -1.207 | -0.6800 | Yes |
| 98 | TP53 | tumor protein p53 [Source:HGNC Symbol;Acc:HGNC:11998] | 34112 | -1.214 | -0.6764 | Yes |
| 99 | TIMELESS | timeless circadian regulator [Source:HGNC Symbol;Acc:HGNC:11813] | 34130 | -1.218 | -0.6727 | Yes |
| 100 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34219 | -1.252 | -0.6709 | Yes |
| 101 | TUBG1 | tubulin gamma 1 [Source:HGNC Symbol;Acc:HGNC:12417] | 34234 | -1.257 | -0.6670 | Yes |
| 102 | EXOSC8 | exosome component 8 [Source:HGNC Symbol;Acc:HGNC:17035] | 34270 | -1.270 | -0.6636 | Yes |
| 103 | USP1 | ubiquitin specific peptidase 1 [Source:HGNC Symbol;Acc:HGNC:12607] | 34285 | -1.275 | -0.6596 | Yes |
| 104 | CDK4 | cyclin dependent kinase 4 [Source:HGNC Symbol;Acc:HGNC:1773] | 34451 | -1.336 | -0.6595 | Yes |
| 105 | PDS5B | PDS5 cohesin associated factor B [Source:HGNC Symbol;Acc:HGNC:20418] | 34480 | -1.345 | -0.6557 | Yes |
| 106 | MCM3 | minichromosome maintenance complex component 3 [Source:HGNC Symbol;Acc:HGNC:6945] | 34522 | -1.361 | -0.6521 | Yes |
| 107 | CDC20 | cell division cycle 20 [Source:HGNC Symbol;Acc:HGNC:1723] | 34533 | -1.364 | -0.6478 | Yes |
| 108 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 34584 | -1.386 | -0.6444 | Yes |
| 109 | MCM4 | minichromosome maintenance complex component 4 [Source:HGNC Symbol;Acc:HGNC:6947] | 34655 | -1.411 | -0.6415 | Yes |
| 110 | RFC1 | replication factor C subunit 1 [Source:HGNC Symbol;Acc:HGNC:9969] | 34702 | -1.428 | -0.6378 | Yes |
| 111 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34705 | -1.428 | -0.6330 | Yes |
| 112 | MRE11 | MRE11 homolog, double strand break repair nuclease [Source:HGNC Symbol;Acc:HGNC:7230] | 34816 | -1.469 | -0.6310 | Yes |
| 113 | KIF2C | kinesin family member 2C [Source:HGNC Symbol;Acc:HGNC:6393] | 34826 | -1.475 | -0.6262 | Yes |
| 114 | PLK1 | polo like kinase 1 [Source:HGNC Symbol;Acc:HGNC:9077] | 34860 | -1.492 | -0.6220 | Yes |
| 115 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 34894 | -1.508 | -0.6178 | Yes |
| 116 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 34896 | -1.510 | -0.6126 | Yes |
| 117 | CIT | citron rho-interacting serine/threonine kinase [Source:HGNC Symbol;Acc:HGNC:1985] | 34904 | -1.517 | -0.6076 | Yes |
| 118 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 34976 | -1.554 | -0.6043 | Yes |
| 119 | PPM1D | protein phosphatase, Mg2+/Mn2+ dependent 1D [Source:HGNC Symbol;Acc:HGNC:9277] | 34984 | -1.558 | -0.5991 | Yes |
| 120 | DIAPH3 | diaphanous related formin 3 [Source:HGNC Symbol;Acc:HGNC:15480] | 34999 | -1.564 | -0.5942 | Yes |
| 121 | ORC6 | origin recognition complex subunit 6 [Source:HGNC Symbol;Acc:HGNC:17151] | 35016 | -1.575 | -0.5892 | Yes |
| 122 | RNASEH2A | ribonuclease H2 subunit A [Source:HGNC Symbol;Acc:HGNC:18518] | 35022 | -1.580 | -0.5840 | Yes |
| 123 | DSCC1 | DNA replication and sister chromatid cohesion 1 [Source:HGNC Symbol;Acc:HGNC:24453] | 35083 | -1.617 | -0.5801 | Yes |
| 124 | NASP | nuclear autoantigenic sperm protein [Source:HGNC Symbol;Acc:HGNC:7644] | 35131 | -1.642 | -0.5758 | Yes |
| 125 | MMS22L | MMS22 like, DNA repair protein [Source:HGNC Symbol;Acc:HGNC:21475] | 35133 | -1.643 | -0.5702 | Yes |
| 126 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 35165 | -1.659 | -0.5654 | Yes |
| 127 | SPAG5 | sperm associated antigen 5 [Source:HGNC Symbol;Acc:HGNC:13452] | 35207 | -1.677 | -0.5607 | Yes |
| 128 | SMC1A | structural maintenance of chromosomes 1A [Source:HGNC Symbol;Acc:HGNC:11111] | 35233 | -1.694 | -0.5556 | Yes |
| 129 | RPA2 | replication protein A2 [Source:HGNC Symbol;Acc:HGNC:10290] | 35234 | -1.694 | -0.5499 | Yes |
| 130 | RAD21 | RAD21 cohesin complex component [Source:HGNC Symbol;Acc:HGNC:9811] | 35250 | -1.704 | -0.5444 | Yes |
| 131 | CDCA8 | cell division cycle associated 8 [Source:HGNC Symbol;Acc:HGNC:14629] | 35266 | -1.714 | -0.5390 | Yes |
| 132 | ASF1B | anti-silencing function 1B histone chaperone [Source:HGNC Symbol;Acc:HGNC:20996] | 35303 | -1.732 | -0.5341 | Yes |
| 133 | CBX5 | chromobox 5 [Source:HGNC Symbol;Acc:HGNC:1555] | 35316 | -1.738 | -0.5285 | Yes |
| 134 | PMS2 | PMS1 homolog 2, mismatch repair system component [Source:HGNC Symbol;Acc:HGNC:9122] | 35327 | -1.743 | -0.5228 | Yes |
| 135 | SPC25 | SPC25 component of NDC80 kinetochore complex [Source:HGNC Symbol;Acc:HGNC:24031] | 35454 | -1.811 | -0.5200 | Yes |
| 136 | DEK | DEK proto-oncogene [Source:HGNC Symbol;Acc:HGNC:2768] | 35509 | -1.842 | -0.5152 | Yes |
| 137 | KIF18B | kinesin family member 18B [Source:HGNC Symbol;Acc:HGNC:27102] | 35530 | -1.855 | -0.5094 | Yes |
| 138 | SMC3 | structural maintenance of chromosomes 3 [Source:HGNC Symbol;Acc:HGNC:2468] | 35567 | -1.877 | -0.5040 | Yes |
| 139 | CCP110 | centriolar coiled-coil protein 110 [Source:HGNC Symbol;Acc:HGNC:24342] | 35634 | -1.925 | -0.4992 | Yes |
| 140 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 35645 | -1.933 | -0.4929 | Yes |
| 141 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 35659 | -1.943 | -0.4866 | Yes |
| 142 | MXD3 | MAX dimerization protein 3 [Source:HGNC Symbol;Acc:HGNC:14008] | 35700 | -1.974 | -0.4809 | Yes |
| 143 | HMMR | hyaluronan mediated motility receptor [Source:HGNC Symbol;Acc:HGNC:5012] | 35723 | -1.996 | -0.4747 | Yes |
| 144 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 35732 | -2.004 | -0.4681 | Yes |
| 145 | UBE2T | ubiquitin conjugating enzyme E2 T [Source:HGNC Symbol;Acc:HGNC:25009] | 35736 | -2.006 | -0.4613 | Yes |
| 146 | HELLS | helicase, lymphoid specific [Source:HGNC Symbol;Acc:HGNC:4861] | 35750 | -2.017 | -0.4548 | Yes |
| 147 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35755 | -2.023 | -0.4480 | Yes |
| 148 | CDKN1B | cyclin dependent kinase inhibitor 1B [Source:HGNC Symbol;Acc:HGNC:1785] | 35764 | -2.028 | -0.4413 | Yes |
| 149 | KIF4A | kinesin family member 4A [Source:HGNC Symbol;Acc:HGNC:13339] | 35793 | -2.055 | -0.4351 | Yes |
| 150 | RAD51C | RAD51 paralog C [Source:HGNC Symbol;Acc:HGNC:9820] | 35799 | -2.059 | -0.4282 | Yes |
| 151 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 35876 | -2.113 | -0.4230 | Yes |
| 152 | BIRC5 | baculoviral IAP repeat containing 5 [Source:HGNC Symbol;Acc:HGNC:593] | 35889 | -2.122 | -0.4161 | Yes |
| 153 | MCM2 | minichromosome maintenance complex component 2 [Source:HGNC Symbol;Acc:HGNC:6944] | 35912 | -2.142 | -0.4094 | Yes |
| 154 | POLE | DNA polymerase epsilon, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9177] | 35916 | -2.145 | -0.4022 | Yes |
| 155 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 35935 | -2.155 | -0.3953 | Yes |
| 156 | TOP2A | DNA topoisomerase II alpha [Source:HGNC Symbol;Acc:HGNC:11989] | 35959 | -2.181 | -0.3885 | Yes |
| 157 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 35963 | -2.182 | -0.3811 | Yes |
| 158 | PSIP1 | PC4 and SFRS1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:9527] | 35969 | -2.187 | -0.3738 | Yes |
| 159 | PRIM2 | DNA primase subunit 2 [Source:HGNC Symbol;Acc:HGNC:9370] | 36007 | -2.209 | -0.3672 | Yes |
| 160 | MCM6 | minichromosome maintenance complex component 6 [Source:HGNC Symbol;Acc:HGNC:6949] | 36012 | -2.211 | -0.3598 | Yes |
| 161 | TUBB | tubulin beta class I [Source:HGNC Symbol;Acc:HGNC:20778] | 36040 | -2.235 | -0.3529 | Yes |
| 162 | CCNB2 | cyclin B2 [Source:HGNC Symbol;Acc:HGNC:1580] | 36081 | -2.263 | -0.3463 | Yes |
| 163 | HMGB3 | high mobility group box 3 [Source:HGNC Symbol;Acc:HGNC:5004] | 36099 | -2.278 | -0.3390 | Yes |
| 164 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 36186 | -2.355 | -0.3333 | Yes |
| 165 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36191 | -2.357 | -0.3253 | Yes |
| 166 | GINS3 | GINS complex subunit 3 [Source:HGNC Symbol;Acc:HGNC:25851] | 36270 | -2.438 | -0.3191 | Yes |
| 167 | UBR7 | ubiquitin protein ligase E3 component n-recognin 7 [Source:HGNC Symbol;Acc:HGNC:20344] | 36275 | -2.443 | -0.3109 | Yes |
| 168 | RPA1 | replication protein A1 [Source:HGNC Symbol;Acc:HGNC:10289] | 36284 | -2.451 | -0.3027 | Yes |
| 169 | AURKB | aurora kinase B [Source:HGNC Symbol;Acc:HGNC:11390] | 36286 | -2.451 | -0.2944 | Yes |
| 170 | MELK | maternal embryonic leucine zipper kinase [Source:HGNC Symbol;Acc:HGNC:16870] | 36289 | -2.458 | -0.2861 | Yes |
| 171 | RACGAP1 | Rac GTPase activating protein 1 [Source:HGNC Symbol;Acc:HGNC:9804] | 36326 | -2.499 | -0.2785 | Yes |
| 172 | BARD1 | BRCA1 associated RING domain 1 [Source:HGNC Symbol;Acc:HGNC:952] | 36327 | -2.502 | -0.2700 | Yes |
| 173 | CENPM | centromere protein M [Source:HGNC Symbol;Acc:HGNC:18352] | 36383 | -2.566 | -0.2627 | Yes |
| 174 | ATAD2 | ATPase family AAA domain containing 2 [Source:HGNC Symbol;Acc:HGNC:30123] | 36387 | -2.570 | -0.2540 | Yes |
| 175 | RFC2 | replication factor C subunit 2 [Source:HGNC Symbol;Acc:HGNC:9970] | 36418 | -2.602 | -0.2460 | Yes |
| 176 | ESPL1 | extra spindle pole bodies like 1, separase [Source:HGNC Symbol;Acc:HGNC:16856] | 36438 | -2.629 | -0.2375 | Yes |
| 177 | CDC25B | cell division cycle 25B [Source:HGNC Symbol;Acc:HGNC:1726] | 36454 | -2.651 | -0.2289 | Yes |
| 178 | WEE1 | WEE1 G2 checkpoint kinase [Source:HGNC Symbol;Acc:HGNC:12761] | 36455 | -2.653 | -0.2198 | Yes |
| 179 | RAD51AP1 | RAD51 associated protein 1 [Source:HGNC Symbol;Acc:HGNC:16956] | 36458 | -2.656 | -0.2108 | Yes |
| 180 | PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase [Source:HGNC Symbol;Acc:HGNC:8587] | 36467 | -2.673 | -0.2019 | Yes |
| 181 | H2AX | H2A.X variant histone [Source:HGNC Symbol;Acc:HGNC:4739] | 36470 | -2.674 | -0.1928 | Yes |
| 182 | GINS1 | GINS complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:28980] | 36505 | -2.720 | -0.1845 | Yes |
| 183 | NCAPD2 | non-SMC condensin I complex subunit D2 [Source:HGNC Symbol;Acc:HGNC:24305] | 36519 | -2.751 | -0.1754 | Yes |
| 184 | MCM7 | minichromosome maintenance complex component 7 [Source:HGNC Symbol;Acc:HGNC:6950] | 36546 | -2.789 | -0.1666 | Yes |
| 185 | CDCA3 | cell division cycle associated 3 [Source:HGNC Symbol;Acc:HGNC:14624] | 36562 | -2.813 | -0.1574 | Yes |
| 186 | BRCA2 | BRCA2 DNA repair associated [Source:HGNC Symbol;Acc:HGNC:1101] | 36585 | -2.864 | -0.1483 | Yes |
| 187 | E2F8 | E2F transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24727] | 36622 | -2.908 | -0.1393 | Yes |
| 188 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36627 | -2.912 | -0.1295 | Yes |
| 189 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 36629 | -2.914 | -0.1196 | Yes |
| 190 | MYBL2 | MYB proto-oncogene like 2 [Source:HGNC Symbol;Acc:HGNC:7548] | 36643 | -2.941 | -0.1099 | Yes |
| 191 | POLD1 | DNA polymerase delta 1, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9175] | 36649 | -2.950 | -0.0999 | Yes |
| 192 | RRM2 | ribonucleotide reductase regulatory subunit M2 [Source:HGNC Symbol;Acc:HGNC:10452] | 36679 | -3.000 | -0.0905 | Yes |
| 193 | DNMT1 | DNA methyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:2976] | 36757 | -3.180 | -0.0817 | Yes |
| 194 | CCNE1 | cyclin E1 [Source:HGNC Symbol;Acc:HGNC:1589] | 36789 | -3.243 | -0.0715 | Yes |
| 195 | BRCA1 | BRCA1 DNA repair associated [Source:HGNC Symbol;Acc:HGNC:1100] | 36830 | -3.352 | -0.0611 | Yes |
| 196 | CHEK2 | checkpoint kinase 2 [Source:HGNC Symbol;Acc:HGNC:16627] | 36870 | -3.488 | -0.0503 | Yes |
| 197 | TCF19 | transcription factor 19 [Source:HGNC Symbol;Acc:HGNC:11629] | 36957 | -3.797 | -0.0397 | Yes |
| 198 | MSH2 | mutS homolog 2 [Source:HGNC Symbol;Acc:HGNC:7325] | 37024 | -4.108 | -0.0274 | Yes |
| 199 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37029 | -4.153 | -0.0134 | Yes |
| 200 | UNG | uracil DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:12572] | 37063 | -4.399 | 0.0007 | Yes |