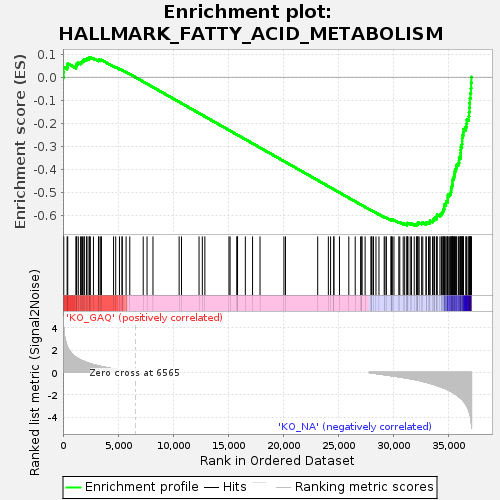
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.6447103 |
| Normalized Enrichment Score (NES) | -1.3999296 |
| Nominal p-value | 0.0010989011 |
| FDR q-value | 0.103582926 |
| FWER p-Value | 0.27 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IL4I1 | interleukin 4 induced 1 [Source:HGNC Symbol;Acc:HGNC:19094] | 55 | 4.029 | 0.0213 | No |
| 2 | ODC1 | ornithine decarboxylase 1 [Source:HGNC Symbol;Acc:HGNC:8109] | 61 | 3.985 | 0.0437 | No |
| 3 | ACSL4 | acyl-CoA synthetase long chain family member 4 [Source:HGNC Symbol;Acc:HGNC:3571] | 375 | 2.305 | 0.0483 | No |
| 4 | CD36 | CD36 molecule [Source:HGNC Symbol;Acc:HGNC:1663] | 431 | 2.203 | 0.0592 | No |
| 5 | HSPH1 | heat shock protein family H (Hsp110) member 1 [Source:HGNC Symbol;Acc:HGNC:16969] | 1182 | 1.340 | 0.0465 | No |
| 6 | ACOX1 | acyl-CoA oxidase 1 [Source:HGNC Symbol;Acc:HGNC:119] | 1199 | 1.330 | 0.0536 | No |
| 7 | UROS | uroporphyrinogen III synthase [Source:HGNC Symbol;Acc:HGNC:12592] | 1244 | 1.303 | 0.0598 | No |
| 8 | UBE2L6 | ubiquitin conjugating enzyme E2 L6 [Source:HGNC Symbol;Acc:HGNC:12490] | 1390 | 1.210 | 0.0627 | No |
| 9 | BLVRA | biliverdin reductase A [Source:HGNC Symbol;Acc:HGNC:1062] | 1594 | 1.119 | 0.0635 | No |
| 10 | HSP90AA1 | heat shock protein 90 alpha family class A member 1 [Source:HGNC Symbol;Acc:HGNC:5253] | 1693 | 1.067 | 0.0669 | No |
| 11 | ME1 | malic enzyme 1 [Source:HGNC Symbol;Acc:HGNC:6983] | 1774 | 1.025 | 0.0705 | No |
| 12 | HSD17B7 | hydroxysteroid 17-beta dehydrogenase 7 [Source:HGNC Symbol;Acc:HGNC:5215] | 1901 | 0.978 | 0.0727 | No |
| 13 | DLD | dihydrolipoamide dehydrogenase [Source:HGNC Symbol;Acc:HGNC:2898] | 1930 | 0.964 | 0.0774 | No |
| 14 | AOC3 | amine oxidase copper containing 3 [Source:HGNC Symbol;Acc:HGNC:550] | 2126 | 0.887 | 0.0771 | No |
| 15 | GABARAPL1 | GABA type A receptor associated protein like 1 [Source:HGNC Symbol;Acc:HGNC:4068] | 2181 | 0.870 | 0.0806 | No |
| 16 | PSME1 | proteasome activator subunit 1 [Source:HGNC Symbol;Acc:HGNC:9568] | 2375 | 0.800 | 0.0799 | No |
| 17 | ADH7 | alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide [Source:HGNC Symbol;Acc:HGNC:256] | 2384 | 0.795 | 0.0841 | No |
| 18 | SERINC1 | serine incorporator 1 [Source:HGNC Symbol;Acc:HGNC:13464] | 2483 | 0.770 | 0.0858 | No |
| 19 | METAP1 | methionyl aminopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:15789] | 2763 | 0.690 | 0.0822 | No |
| 20 | FASN | fatty acid synthase [Source:HGNC Symbol;Acc:HGNC:3594] | 3219 | 0.579 | 0.0731 | No |
| 21 | IDI1 | isopentenyl-diphosphate delta isomerase 1 [Source:HGNC Symbol;Acc:HGNC:5387] | 3265 | 0.568 | 0.0751 | No |
| 22 | OSTC | oligosaccharyltransferase complex non-catalytic subunit [Source:HGNC Symbol;Acc:HGNC:24448] | 3421 | 0.532 | 0.0739 | No |
| 23 | YWHAH | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein eta [Source:HGNC Symbol;Acc:HGNC:12853] | 3491 | 0.516 | 0.0750 | No |
| 24 | PTPRG | protein tyrosine phosphatase receptor type G [Source:HGNC Symbol;Acc:HGNC:9671] | 4589 | 0.312 | 0.0471 | No |
| 25 | SMS | spermine synthase [Source:HGNC Symbol;Acc:HGNC:11123] | 4785 | 0.281 | 0.0434 | No |
| 26 | SLC22A5 | solute carrier family 22 member 5 [Source:HGNC Symbol;Acc:HGNC:10969] | 5125 | 0.219 | 0.0354 | No |
| 27 | ADSL | adenylosuccinate lyase [Source:HGNC Symbol;Acc:HGNC:291] | 5358 | 0.182 | 0.0302 | No |
| 28 | SDHC | succinate dehydrogenase complex subunit C [Source:HGNC Symbol;Acc:HGNC:10682] | 5376 | 0.179 | 0.0307 | No |
| 29 | MDH2 | malate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:6971] | 5728 | 0.134 | 0.0220 | No |
| 30 | ADIPOR2 | adiponectin receptor 2 [Source:HGNC Symbol;Acc:HGNC:24041] | 6063 | 0.085 | 0.0134 | No |
| 31 | AQP7 | aquaporin 7 [Source:HGNC Symbol;Acc:HGNC:640] | 7281 | 0.000 | -0.0195 | No |
| 32 | PCBD1 | pterin-4 alpha-carbinolamine dehydratase 1 [Source:HGNC Symbol;Acc:HGNC:8646] | 7629 | 0.000 | -0.0289 | No |
| 33 | EHHADH | enoyl-CoA hydratase and 3-hydroxyacyl CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3247] | 8168 | 0.000 | -0.0435 | No |
| 34 | AADAT | aminoadipate aminotransferase [Source:HGNC Symbol;Acc:HGNC:17929] | 10542 | 0.000 | -0.1077 | No |
| 35 | CYP1A1 | cytochrome P450 family 1 subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:2595] | 10758 | 0.000 | -0.1136 | No |
| 36 | DECR1 | 2,4-dienoyl-CoA reductase 1 [Source:HGNC Symbol;Acc:HGNC:2753] | 12350 | 0.000 | -0.1566 | No |
| 37 | GAD2 | glutamate decarboxylase 2 [Source:HGNC Symbol;Acc:HGNC:4093] | 12648 | 0.000 | -0.1647 | No |
| 38 | VNN1 | vanin 1 [Source:HGNC Symbol;Acc:HGNC:12705] | 12879 | 0.000 | -0.1709 | No |
| 39 | INMT | indolethylamine N-methyltransferase [Source:HGNC Symbol;Acc:HGNC:6069] | 15065 | 0.000 | -0.2301 | No |
| 40 | CIDEA | cell death inducing DFFA like effector a [Source:HGNC Symbol;Acc:HGNC:1976] | 15166 | 0.000 | -0.2328 | No |
| 41 | CA4 | carbonic anhydrase 4 [Source:HGNC Symbol;Acc:HGNC:1375] | 15794 | 0.000 | -0.2498 | No |
| 42 | CA6 | carbonic anhydrase 6 [Source:HGNC Symbol;Acc:HGNC:1380] | 15796 | 0.000 | -0.2498 | No |
| 43 | CEL | carboxyl ester lipase [Source:HGNC Symbol;Acc:HGNC:1848] | 15845 | 0.000 | -0.2511 | No |
| 44 | GAPDHS | glyceraldehyde-3-phosphate dehydrogenase, spermatogenic [Source:HGNC Symbol;Acc:HGNC:24864] | 16552 | 0.000 | -0.2702 | No |
| 45 | MGLL | monoglyceride lipase [Source:HGNC Symbol;Acc:HGNC:17038] | 17208 | 0.000 | -0.2879 | No |
| 46 | XIST | X inactive specific transcript [Source:HGNC Symbol;Acc:HGNC:12810] | 17886 | 0.000 | -0.3063 | No |
| 47 | HMGCS2 | 3-hydroxy-3-methylglutaryl-CoA synthase 2 [Source:HGNC Symbol;Acc:HGNC:5008] | 20055 | 0.000 | -0.3650 | No |
| 48 | TDO2 | tryptophan 2,3-dioxygenase [Source:HGNC Symbol;Acc:HGNC:11708] | 20197 | 0.000 | -0.3688 | No |
| 49 | CD1D | CD1d molecule [Source:HGNC Symbol;Acc:HGNC:1637] | 23121 | 0.000 | -0.4479 | No |
| 50 | ACSM3 | acyl-CoA synthetase medium chain family member 3 [Source:HGNC Symbol;Acc:HGNC:10522] | 24089 | 0.000 | -0.4741 | No |
| 51 | PPARA | peroxisome proliferator activated receptor alpha [Source:HGNC Symbol;Acc:HGNC:9232] | 24292 | 0.000 | -0.4796 | No |
| 52 | CYP4A22 | cytochrome P450 family 4 subfamily A member 22 [Source:HGNC Symbol;Acc:HGNC:20575] | 24581 | 0.000 | -0.4874 | No |
| 53 | CYP4A11 | cytochrome P450 family 4 subfamily A member 11 [Source:HGNC Symbol;Acc:HGNC:2642] | 24592 | 0.000 | -0.4877 | No |
| 54 | HAO2 | hydroxyacid oxidase 2 [Source:HGNC Symbol;Acc:HGNC:4810] | 25097 | 0.000 | -0.5013 | No |
| 55 | ALDH1A1 | aldehyde dehydrogenase 1 family member A1 [Source:HGNC Symbol;Acc:HGNC:402] | 25945 | 0.000 | -0.5242 | No |
| 56 | ALDH3A1 | aldehyde dehydrogenase 3 family member A1 [Source:HGNC Symbol;Acc:HGNC:405] | 26527 | 0.000 | -0.5400 | No |
| 57 | ADH1C | alcohol dehydrogenase 1C (class I), gamma polypeptide [Source:HGNC Symbol;Acc:HGNC:251] | 27016 | 0.000 | -0.5532 | No |
| 58 | FABP1 | fatty acid binding protein 1 [Source:HGNC Symbol;Acc:HGNC:3555] | 27132 | 0.000 | -0.5563 | No |
| 59 | FABP2 | fatty acid binding protein 2 [Source:HGNC Symbol;Acc:HGNC:3556] | 27133 | 0.000 | -0.5563 | No |
| 60 | FMO1 | flavin containing dimethylaniline monoxygenase 1 [Source:HGNC Symbol;Acc:HGNC:3769] | 27426 | 0.000 | -0.5642 | No |
| 61 | HCCS | holocytochrome c synthase [Source:HGNC Symbol;Acc:HGNC:4837] | 27950 | -0.029 | -0.5782 | No |
| 62 | SUCLG1 | succinate-CoA ligase GDP/ADP-forming subunit alpha [Source:HGNC Symbol;Acc:HGNC:11449] | 28028 | -0.036 | -0.5801 | No |
| 63 | ACADVL | acyl-CoA dehydrogenase very long chain [Source:HGNC Symbol;Acc:HGNC:92] | 28040 | -0.037 | -0.5802 | No |
| 64 | BMPR1B | bone morphogenetic protein receptor type 1B [Source:HGNC Symbol;Acc:HGNC:1077] | 28186 | -0.061 | -0.5837 | No |
| 65 | PDHB | pyruvate dehydrogenase E1 subunit beta [Source:HGNC Symbol;Acc:HGNC:8808] | 28410 | -0.091 | -0.5893 | No |
| 66 | NBN | nibrin [Source:HGNC Symbol;Acc:HGNC:7652] | 28683 | -0.130 | -0.5959 | No |
| 67 | CA2 | carbonic anhydrase 2 [Source:HGNC Symbol;Acc:HGNC:1373] | 29157 | -0.193 | -0.6076 | No |
| 68 | RDH11 | retinol dehydrogenase 11 [Source:HGNC Symbol;Acc:HGNC:17964] | 29229 | -0.204 | -0.6084 | No |
| 69 | S100A10 | S100 calcium binding protein A10 [Source:HGNC Symbol;Acc:HGNC:10487] | 29360 | -0.226 | -0.6106 | No |
| 70 | G0S2 | G0/G1 switch 2 [Source:HGNC Symbol;Acc:HGNC:30229] | 29778 | -0.287 | -0.6203 | No |
| 71 | HSD17B11 | hydroxysteroid 17-beta dehydrogenase 11 [Source:HGNC Symbol;Acc:HGNC:22960] | 29798 | -0.290 | -0.6192 | No |
| 72 | DLST | dihydrolipoamide S-succinyltransferase [Source:HGNC Symbol;Acc:HGNC:2911] | 29812 | -0.291 | -0.6179 | No |
| 73 | GPD2 | glycerol-3-phosphate dehydrogenase 2 [Source:HGNC Symbol;Acc:HGNC:4456] | 29896 | -0.304 | -0.6184 | No |
| 74 | HMGCS1 | 3-hydroxy-3-methylglutaryl-CoA synthase 1 [Source:HGNC Symbol;Acc:HGNC:5007] | 30044 | -0.322 | -0.6206 | No |
| 75 | KMT5A | lysine methyltransferase 5A [Source:HGNC Symbol;Acc:HGNC:29489] | 30486 | -0.381 | -0.6303 | No |
| 76 | APEX1 | apurinic/apyrimidinic endodeoxyribonuclease 1 [Source:HGNC Symbol;Acc:HGNC:587] | 30574 | -0.396 | -0.6305 | No |
| 77 | GPD1 | glycerol-3-phosphate dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:4455] | 30883 | -0.447 | -0.6363 | No |
| 78 | ACO2 | aconitase 2 [Source:HGNC Symbol;Acc:HGNC:118] | 31018 | -0.465 | -0.6373 | No |
| 79 | RDH16 | retinol dehydrogenase 16 [Source:HGNC Symbol;Acc:HGNC:29674] | 31218 | -0.499 | -0.6398 | No |
| 80 | TP53INP2 | tumor protein p53 inducible nuclear protein 2 [Source:HGNC Symbol;Acc:HGNC:16104] | 31226 | -0.500 | -0.6372 | No |
| 81 | ENO3 | enolase 3 [Source:HGNC Symbol;Acc:HGNC:3354] | 31272 | -0.506 | -0.6356 | No |
| 82 | SUCLA2 | succinate-CoA ligase ADP-forming subunit beta [Source:HGNC Symbol;Acc:HGNC:11448] | 31317 | -0.513 | -0.6339 | No |
| 83 | ACSL5 | acyl-CoA synthetase long chain family member 5 [Source:HGNC Symbol;Acc:HGNC:16526] | 31518 | -0.549 | -0.6362 | No |
| 84 | MAOA | monoamine oxidase A [Source:HGNC Symbol;Acc:HGNC:6833] | 31634 | -0.567 | -0.6361 | No |
| 85 | ALDH3A2 | aldehyde dehydrogenase 3 family member A2 [Source:HGNC Symbol;Acc:HGNC:403] | 31882 | -0.613 | -0.6393 | No |
| 86 | ACSS1 | acyl-CoA synthetase short chain family member 1 [Source:HGNC Symbol;Acc:HGNC:16091] | 32083 | -0.659 | -0.6410 | Yes |
| 87 | FH | fumarate hydratase [Source:HGNC Symbol;Acc:HGNC:3700] | 32136 | -0.669 | -0.6386 | Yes |
| 88 | IDH3B | isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit beta [Source:HGNC Symbol;Acc:HGNC:5385] | 32208 | -0.684 | -0.6367 | Yes |
| 89 | ECI1 | enoyl-CoA delta isomerase 1 [Source:HGNC Symbol;Acc:HGNC:2703] | 32219 | -0.687 | -0.6331 | Yes |
| 90 | ACOT8 | acyl-CoA thioesterase 8 [Source:HGNC Symbol;Acc:HGNC:15919] | 32328 | -0.708 | -0.6320 | Yes |
| 91 | PTS | 6-pyruvoyltetrahydropterin synthase [Source:HGNC Symbol;Acc:HGNC:9689] | 32557 | -0.759 | -0.6338 | Yes |
| 92 | PDHA1 | pyruvate dehydrogenase E1 subunit alpha 1 [Source:HGNC Symbol;Acc:HGNC:8806] | 32661 | -0.787 | -0.6322 | Yes |
| 93 | HIBCH | 3-hydroxyisobutyryl-CoA hydrolase [Source:HGNC Symbol;Acc:HGNC:4908] | 32961 | -0.864 | -0.6354 | Yes |
| 94 | HSDL2 | hydroxysteroid dehydrogenase like 2 [Source:HGNC Symbol;Acc:HGNC:18572] | 32967 | -0.865 | -0.6306 | Yes |
| 95 | MIF | macrophage migration inhibitory factor [Source:HGNC Symbol;Acc:HGNC:7097] | 33182 | -0.922 | -0.6312 | Yes |
| 96 | SDHD | succinate dehydrogenase complex subunit D [Source:HGNC Symbol;Acc:HGNC:10683] | 33300 | -0.958 | -0.6290 | Yes |
| 97 | LDHA | lactate dehydrogenase A [Source:HGNC Symbol;Acc:HGNC:6535] | 33313 | -0.961 | -0.6239 | Yes |
| 98 | CPOX | coproporphyrinogen oxidase [Source:HGNC Symbol;Acc:HGNC:2321] | 33546 | -1.034 | -0.6243 | Yes |
| 99 | NSDHL | NAD(P) dependent steroid dehydrogenase-like [Source:HGNC Symbol;Acc:HGNC:13398] | 33600 | -1.049 | -0.6198 | Yes |
| 100 | ACSL1 | acyl-CoA synthetase long chain family member 1 [Source:HGNC Symbol;Acc:HGNC:3569] | 33710 | -1.081 | -0.6166 | Yes |
| 101 | HADHB | hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit beta [Source:HGNC Symbol;Acc:HGNC:4803] | 33740 | -1.089 | -0.6112 | Yes |
| 102 | SUCLG2 | succinate-CoA ligase GDP-forming subunit beta [Source:HGNC Symbol;Acc:HGNC:11450] | 33928 | -1.159 | -0.6098 | Yes |
| 103 | HMGCL | 3-hydroxy-3-methylglutaryl-CoA lyase [Source:HGNC Symbol;Acc:HGNC:5005] | 33935 | -1.162 | -0.6033 | Yes |
| 104 | ETFDH | electron transfer flavoprotein dehydrogenase [Source:HGNC Symbol;Acc:HGNC:3483] | 33959 | -1.171 | -0.5973 | Yes |
| 105 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34219 | -1.252 | -0.5973 | Yes |
| 106 | PRDX6 | peroxiredoxin 6 [Source:HGNC Symbol;Acc:HGNC:16753] | 34349 | -1.299 | -0.5934 | Yes |
| 107 | IDH3G | isocitrate dehydrogenase (NAD(+)) 3 non-catalytic subunit gamma [Source:HGNC Symbol;Acc:HGNC:5386] | 34393 | -1.316 | -0.5871 | Yes |
| 108 | RAP1GDS1 | Rap1 GTPase-GDP dissociation stimulator 1 [Source:HGNC Symbol;Acc:HGNC:9859] | 34488 | -1.348 | -0.5821 | Yes |
| 109 | BCKDHB | branched chain keto acid dehydrogenase E1 subunit beta [Source:HGNC Symbol;Acc:HGNC:987] | 34527 | -1.362 | -0.5754 | Yes |
| 110 | CCDC58 | coiled-coil domain containing 58 [Source:HGNC Symbol;Acc:HGNC:31136] | 34595 | -1.389 | -0.5693 | Yes |
| 111 | SDHA | succinate dehydrogenase complex flavoprotein subunit A [Source:HGNC Symbol;Acc:HGNC:10680] | 34601 | -1.391 | -0.5616 | Yes |
| 112 | ACADL | acyl-CoA dehydrogenase long chain [Source:HGNC Symbol;Acc:HGNC:88] | 34617 | -1.399 | -0.5541 | Yes |
| 113 | ACAT2 | acetyl-CoA acetyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:94] | 34759 | -1.446 | -0.5497 | Yes |
| 114 | CBR3 | carbonyl reductase 3 [Source:HGNC Symbol;Acc:HGNC:1549] | 34775 | -1.452 | -0.5419 | Yes |
| 115 | LGALS1 | galectin 1 [Source:HGNC Symbol;Acc:HGNC:6561] | 34905 | -1.518 | -0.5368 | Yes |
| 116 | ECHS1 | enoyl-CoA hydratase, short chain 1 [Source:HGNC Symbol;Acc:HGNC:3151] | 34933 | -1.531 | -0.5289 | Yes |
| 117 | MLYCD | malonyl-CoA decarboxylase [Source:HGNC Symbol;Acc:HGNC:7150] | 34936 | -1.532 | -0.5203 | Yes |
| 118 | GSTZ1 | glutathione S-transferase zeta 1 [Source:HGNC Symbol;Acc:HGNC:4643] | 34943 | -1.538 | -0.5118 | Yes |
| 119 | RETSAT | retinol saturase [Source:HGNC Symbol;Acc:HGNC:25991] | 35100 | -1.626 | -0.5068 | Yes |
| 120 | ACADM | acyl-CoA dehydrogenase medium chain [Source:HGNC Symbol;Acc:HGNC:89] | 35197 | -1.671 | -0.4999 | Yes |
| 121 | ALDOA | aldolase, fructose-bisphosphate A [Source:HGNC Symbol;Acc:HGNC:414] | 35228 | -1.689 | -0.4912 | Yes |
| 122 | MDH1 | malate dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:6970] | 35236 | -1.695 | -0.4818 | Yes |
| 123 | CBR1 | carbonyl reductase 1 [Source:HGNC Symbol;Acc:HGNC:1548] | 35295 | -1.728 | -0.4736 | Yes |
| 124 | UGDH | UDP-glucose 6-dehydrogenase [Source:HGNC Symbol;Acc:HGNC:12525] | 35344 | -1.750 | -0.4650 | Yes |
| 125 | D2HGDH | D-2-hydroxyglutarate dehydrogenase [Source:HGNC Symbol;Acc:HGNC:28358] | 35352 | -1.753 | -0.4553 | Yes |
| 126 | ALAD | aminolevulinate dehydratase [Source:HGNC Symbol;Acc:HGNC:395] | 35353 | -1.753 | -0.4454 | Yes |
| 127 | CPT2 | carnitine palmitoyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:2330] | 35443 | -1.805 | -0.4376 | Yes |
| 128 | HSD17B4 | hydroxysteroid 17-beta dehydrogenase 4 [Source:HGNC Symbol;Acc:HGNC:5213] | 35498 | -1.835 | -0.4286 | Yes |
| 129 | UROD | uroporphyrinogen decarboxylase [Source:HGNC Symbol;Acc:HGNC:12591] | 35549 | -1.867 | -0.4194 | Yes |
| 130 | DHCR24 | 24-dehydrocholesterol reductase [Source:HGNC Symbol;Acc:HGNC:2859] | 35555 | -1.870 | -0.4090 | Yes |
| 131 | BPHL | biphenyl hydrolase like [Source:HGNC Symbol;Acc:HGNC:1094] | 35624 | -1.921 | -0.4000 | Yes |
| 132 | MCEE | methylmalonyl-CoA epimerase [Source:HGNC Symbol;Acc:HGNC:16732] | 35693 | -1.967 | -0.3907 | Yes |
| 133 | EPHX1 | epoxide hydrolase 1 [Source:HGNC Symbol;Acc:HGNC:3401] | 35734 | -2.006 | -0.3804 | Yes |
| 134 | ACADS | acyl-CoA dehydrogenase short chain [Source:HGNC Symbol;Acc:HGNC:90] | 35888 | -2.122 | -0.3726 | Yes |
| 135 | ECI2 | enoyl-CoA delta isomerase 2 [Source:HGNC Symbol;Acc:HGNC:14601] | 35952 | -2.174 | -0.3620 | Yes |
| 136 | AUH | AU RNA binding methylglutaconyl-CoA hydratase [Source:HGNC Symbol;Acc:HGNC:890] | 35958 | -2.180 | -0.3498 | Yes |
| 137 | HSD17B10 | hydroxysteroid 17-beta dehydrogenase 10 [Source:HGNC Symbol;Acc:HGNC:4800] | 36094 | -2.272 | -0.3406 | Yes |
| 138 | ENO2 | enolase 2 [Source:HGNC Symbol;Acc:HGNC:3353] | 36098 | -2.276 | -0.3278 | Yes |
| 139 | GCDH | glutaryl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4189] | 36111 | -2.286 | -0.3152 | Yes |
| 140 | NTHL1 | nth like DNA glycosylase 1 [Source:HGNC Symbol;Acc:HGNC:8028] | 36122 | -2.293 | -0.3025 | Yes |
| 141 | ACAA1 | acetyl-CoA acyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:82] | 36210 | -2.370 | -0.2914 | Yes |
| 142 | ELOVL5 | ELOVL fatty acid elongase 5 [Source:HGNC Symbol;Acc:HGNC:21308] | 36230 | -2.394 | -0.2784 | Yes |
| 143 | CRAT | carnitine O-acetyltransferase [Source:HGNC Symbol;Acc:HGNC:2342] | 36234 | -2.397 | -0.2649 | Yes |
| 144 | CRYZ | crystallin zeta [Source:HGNC Symbol;Acc:HGNC:2419] | 36259 | -2.425 | -0.2519 | Yes |
| 145 | ACAA2 | acetyl-CoA acyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:83] | 36347 | -2.523 | -0.2399 | Yes |
| 146 | NCAPH2 | non-SMC condensin II complex subunit H2 [Source:HGNC Symbol;Acc:HGNC:25071] | 36357 | -2.535 | -0.2259 | Yes |
| 147 | ALDH9A1 | aldehyde dehydrogenase 9 family member A1 [Source:HGNC Symbol;Acc:HGNC:412] | 36561 | -2.810 | -0.2155 | Yes |
| 148 | ECH1 | enoyl-CoA hydratase 1 [Source:HGNC Symbol;Acc:HGNC:3149] | 36642 | -2.940 | -0.2010 | Yes |
| 149 | HADH | hydroxyacyl-CoA dehydrogenase [Source:HGNC Symbol;Acc:HGNC:4799] | 36664 | -2.967 | -0.1848 | Yes |
| 150 | ERP29 | endoplasmic reticulum protein 29 [Source:HGNC Symbol;Acc:HGNC:13799] | 36834 | -3.368 | -0.1703 | Yes |
| 151 | ACOT2 | acyl-CoA thioesterase 2 [Source:HGNC Symbol;Acc:HGNC:18431] | 36863 | -3.472 | -0.1514 | Yes |
| 152 | GRHPR | glyoxylate and hydroxypyruvate reductase [Source:HGNC Symbol;Acc:HGNC:4570] | 36900 | -3.572 | -0.1322 | Yes |
| 153 | IDH1 | isocitrate dehydrogenase (NADP(+)) 1 [Source:HGNC Symbol;Acc:HGNC:5382] | 36914 | -3.618 | -0.1121 | Yes |
| 154 | CPT1A | carnitine palmitoyltransferase 1A [Source:HGNC Symbol;Acc:HGNC:2328] | 36933 | -3.693 | -0.0917 | Yes |
| 155 | GLUL | glutamate-ammonia ligase [Source:HGNC Symbol;Acc:HGNC:4341] | 36991 | -3.922 | -0.0710 | Yes |
| 156 | REEP6 | receptor accessory protein 6 [Source:HGNC Symbol;Acc:HGNC:30078] | 37028 | -4.148 | -0.0486 | Yes |
| 157 | LTC4S | leukotriene C4 synthase [Source:HGNC Symbol;Acc:HGNC:6719] | 37051 | -4.307 | -0.0248 | Yes |
| 158 | HPGD | 15-hydroxyprostaglandin dehydrogenase [Source:HGNC Symbol;Acc:HGNC:5154] | 37075 | -4.565 | 0.0004 | Yes |