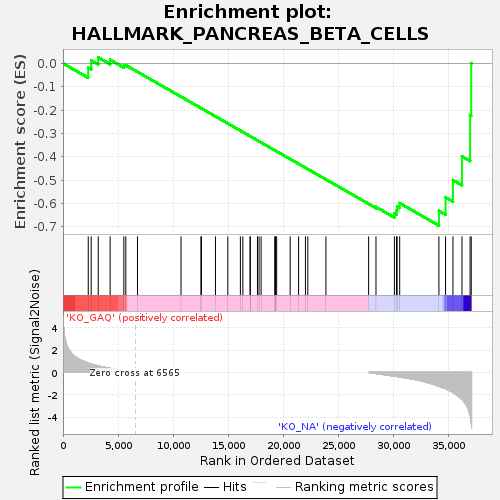
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_GAQ_versus_KO_NA.Nof1.cls #KO_GAQ_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_GAQ_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | HALLMARK_PANCREAS_BETA_CELLS |
| Enrichment Score (ES) | -0.69515824 |
| Normalized Enrichment Score (NES) | -1.3145088 |
| Nominal p-value | 0.064986736 |
| FDR q-value | 0.15047242 |
| FWER p-Value | 0.704 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SRPRB | SRP receptor subunit beta [Source:HGNC Symbol;Acc:HGNC:24085] | 2291 | 0.829 | -0.0189 | No |
| 2 | SRP14 | signal recognition particle 14 [Source:HGNC Symbol;Acc:HGNC:11299] | 2559 | 0.745 | 0.0125 | No |
| 3 | SEC11A | SEC11 homolog A, signal peptidase complex subunit [Source:HGNC Symbol;Acc:HGNC:17718] | 3200 | 0.584 | 0.0255 | No |
| 4 | SPCS1 | signal peptidase complex subunit 1 [Source:HGNC Symbol;Acc:HGNC:23401] | 4278 | 0.366 | 0.0154 | No |
| 5 | PAK3 | p21 (RAC1) activated kinase 3 [Source:HGNC Symbol;Acc:HGNC:8592] | 5522 | 0.158 | -0.0100 | No |
| 6 | PCSK1 | proprotein convertase subtilisin/kexin type 1 [Source:HGNC Symbol;Acc:HGNC:8743] | 5703 | 0.136 | -0.0078 | No |
| 7 | SCGN | secretagogin, EF-hand calcium binding protein [Source:HGNC Symbol;Acc:HGNC:16941] | 6762 | 0.000 | -0.0364 | No |
| 8 | CHGA | chromogranin A [Source:HGNC Symbol;Acc:HGNC:1929] | 10712 | 0.000 | -0.1429 | No |
| 9 | G6PC2 | glucose-6-phosphatase catalytic subunit 2 [Source:HGNC Symbol;Acc:HGNC:28906] | 12519 | 0.000 | -0.1917 | No |
| 10 | SST | somatostatin [Source:HGNC Symbol;Acc:HGNC:11329] | 12569 | 0.000 | -0.1930 | No |
| 11 | SYT13 | synaptotagmin 13 [Source:HGNC Symbol;Acc:HGNC:14962] | 13839 | 0.000 | -0.2273 | No |
| 12 | PAX4 | paired box 4 [Source:HGNC Symbol;Acc:HGNC:8618] | 14962 | 0.000 | -0.2575 | No |
| 13 | DCX | doublecortin [Source:HGNC Symbol;Acc:HGNC:2714] | 16102 | 0.000 | -0.2883 | No |
| 14 | PCSK2 | proprotein convertase subtilisin/kexin type 2 [Source:HGNC Symbol;Acc:HGNC:8744] | 16328 | 0.000 | -0.2943 | No |
| 15 | GCG | glucagon [Source:HGNC Symbol;Acc:HGNC:4191] | 16991 | 0.000 | -0.3122 | No |
| 16 | GCK | glucokinase [Source:HGNC Symbol;Acc:HGNC:4195] | 16993 | 0.000 | -0.3122 | No |
| 17 | HNF1A | HNF1 homeobox A [Source:HGNC Symbol;Acc:HGNC:11621] | 17654 | 0.000 | -0.3301 | No |
| 18 | INS | insulin [Source:HGNC Symbol;Acc:HGNC:6081] | 17797 | 0.000 | -0.3339 | No |
| 19 | DPP4 | dipeptidyl peptidase 4 [Source:HGNC Symbol;Acc:HGNC:3009] | 17983 | 0.000 | -0.3389 | No |
| 20 | NKX2-2 | NK2 homeobox 2 [Source:HGNC Symbol;Acc:HGNC:7835] | 19227 | 0.000 | -0.3724 | No |
| 21 | IAPP | islet amyloid polypeptide [Source:HGNC Symbol;Acc:HGNC:5329] | 19275 | 0.000 | -0.3737 | No |
| 22 | NEUROD1 | neuronal differentiation 1 [Source:HGNC Symbol;Acc:HGNC:7762] | 19281 | 0.000 | -0.3738 | No |
| 23 | NEUROG3 | neurogenin 3 [Source:HGNC Symbol;Acc:HGNC:13806] | 19396 | 0.000 | -0.3769 | No |
| 24 | NKX6-1 | NK6 homeobox 1 [Source:HGNC Symbol;Acc:HGNC:7839] | 20621 | 0.000 | -0.4099 | No |
| 25 | PDX1 | pancreatic and duodenal homeobox 1 [Source:HGNC Symbol;Acc:HGNC:6107] | 21386 | 0.000 | -0.4306 | No |
| 26 | INSM1 | INSM transcriptional repressor 1 [Source:HGNC Symbol;Acc:HGNC:6090] | 22009 | 0.000 | -0.4474 | No |
| 27 | ISL1 | ISL LIM homeobox 1 [Source:HGNC Symbol;Acc:HGNC:6132] | 22225 | 0.000 | -0.4532 | No |
| 28 | FOXA2 | forkhead box A2 [Source:HGNC Symbol;Acc:HGNC:5022] | 23869 | 0.000 | -0.4975 | No |
| 29 | SLC2A2 | solute carrier family 2 member 2 [Source:HGNC Symbol;Acc:HGNC:11006] | 27744 | 0.000 | -0.6021 | No |
| 30 | STXBP1 | syntaxin binding protein 1 [Source:HGNC Symbol;Acc:HGNC:11444] | 28412 | -0.091 | -0.6153 | No |
| 31 | ABCC8 | ATP binding cassette subfamily C member 8 [Source:HGNC Symbol;Acc:HGNC:59] | 30086 | -0.328 | -0.6435 | Yes |
| 32 | SRP9 | signal recognition particle 9 [Source:HGNC Symbol;Acc:HGNC:11304] | 30275 | -0.351 | -0.6304 | Yes |
| 33 | ELP4 | elongator acetyltransferase complex subunit 4 [Source:HGNC Symbol;Acc:HGNC:1171] | 30331 | -0.359 | -0.6133 | Yes |
| 34 | VDR | vitamin D receptor [Source:HGNC Symbol;Acc:HGNC:12679] | 30565 | -0.395 | -0.5991 | Yes |
| 35 | AKT3 | AKT serine/threonine kinase 3 [Source:HGNC Symbol;Acc:HGNC:393] | 34125 | -1.217 | -0.6321 | Yes |
| 36 | PKLR | pyruvate kinase L/R [Source:HGNC Symbol;Acc:HGNC:9020] | 34728 | -1.436 | -0.5740 | Yes |
| 37 | MAFB | MAF bZIP transcription factor B [Source:HGNC Symbol;Acc:HGNC:6408] | 35400 | -1.774 | -0.5001 | Yes |
| 38 | LMO2 | LIM domain only 2 [Source:HGNC Symbol;Acc:HGNC:6642] | 36219 | -2.386 | -0.3986 | Yes |
| 39 | PAX6 | paired box 6 [Source:HGNC Symbol;Acc:HGNC:8620] | 36954 | -3.792 | -0.2219 | Yes |
| 40 | FOXO1 | forkhead box O1 [Source:HGNC Symbol;Acc:HGNC:3819] | 37060 | -4.352 | 0.0008 | Yes |