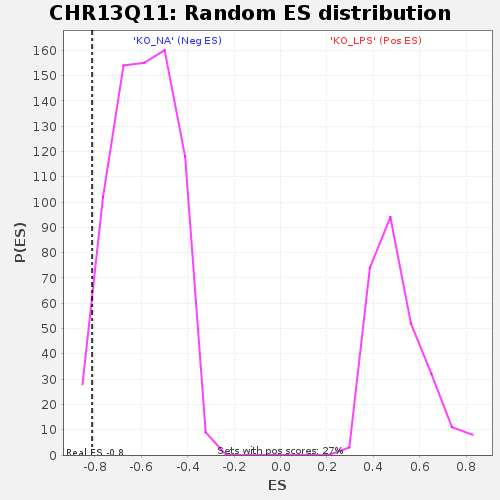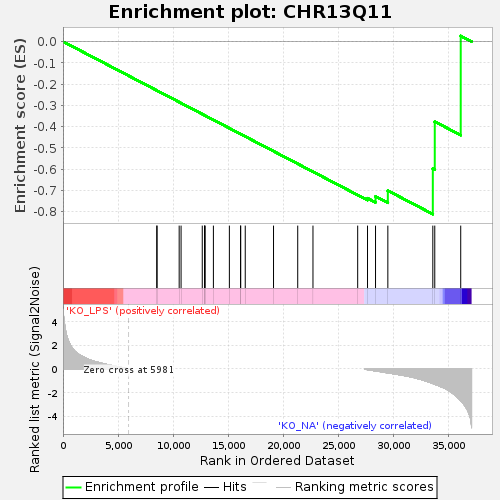
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR13Q11 |
| Enrichment Score (ES) | -0.8107919 |
| Normalized Enrichment Score (NES) | -1.3777981 |
| Nominal p-value | 0.037190083 |
| FDR q-value | 0.47023058 |
| FWER p-Value | 0.963 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ZNF965P | zinc finger protein 965, pseudogene [Source:HGNC Symbol;Acc:HGNC:42025] | 8512 | 0.000 | -0.2296 | No |
| 2 | ZNF962P | zinc finger protein 962, pseudogene [Source:HGNC Symbol;Acc:HGNC:39250] | 8539 | 0.000 | -0.2303 | No |
| 3 | GXYLT1P1 | GXYLT1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:39676] | 10546 | 0.000 | -0.2845 | No |
| 4 | ANKRD20A9P | ankyrin repeat domain 20 family member A9, pseudogene [Source:HGNC Symbol;Acc:HGNC:42023] | 10719 | 0.000 | -0.2891 | No |
| 5 | LINC00328-2P | long intergenic non-protein coding RNA 328-2, pseudogene [Source:HGNC Symbol;Acc:HGNC:42027] | 12642 | 0.000 | -0.3409 | No |
| 6 | LINC00387 | long intergenic non-protein coding RNA 387 [Source:HGNC Symbol;Acc:HGNC:42715] | 12848 | 0.000 | -0.3465 | No |
| 7 | LINC00388 | long intergenic non-protein coding RNA 388 [Source:HGNC Symbol;Acc:HGNC:42716] | 12849 | 0.000 | -0.3465 | No |
| 8 | LINC00349 | long intergenic non-protein coding RNA 349 [Source:HGNC Symbol;Acc:HGNC:42667] | 12906 | 0.000 | -0.3480 | No |
| 9 | SNX18P26 | sorting nexin 18 pseudogene 26 [Source:HGNC Symbol;Acc:HGNC:39634] | 13653 | 0.000 | -0.3681 | No |
| 10 | RNU6-76P | RNA, U6 small nuclear 76, pseudogene [Source:HGNC Symbol;Acc:HGNC:42566] | 15103 | 0.000 | -0.4072 | No |
| 11 | GGT4P | gamma-glutamyltransferase 4 pseudogene [Source:HGNC Symbol;Acc:HGNC:33207] | 16124 | 0.000 | -0.4347 | No |
| 12 | LONRF2P2 | LONRF2 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:39829] | 16126 | 0.000 | -0.4347 | No |
| 13 | BNIP3P7 | BCL2 interacting protein 3 pseudogene 7 [Source:HGNC Symbol;Acc:HGNC:49101] | 16544 | 0.000 | -0.4460 | No |
| 14 | FAM230C | family with sequence similarity 230 member C [Source:HGNC Symbol;Acc:HGNC:24482] | 19111 | 0.000 | -0.5152 | No |
| 15 | RNU6-55P | RNA, U6 small nuclear 55, pseudogene [Source:HGNC Symbol;Acc:HGNC:42545] | 21308 | 0.000 | -0.5745 | No |
| 16 | CYP4F34P | cytochrome P450 family 4 subfamily F member 34, pseudogene [Source:HGNC Symbol;Acc:HGNC:39953] | 22692 | 0.000 | -0.6118 | No |
| 17 | CNN2P12 | calponin 2 pseudogene 12 [Source:HGNC Symbol;Acc:HGNC:39834] | 26751 | 0.000 | -0.7212 | No |
| 18 | FAM207BP | family with sequence similarity 207 member B, pseudogene [Source:HGNC Symbol;Acc:HGNC:39715] | 27644 | -0.052 | -0.7362 | Yes |
| 19 | FEM1AP4 | fem-1 homolog A pseudogene 4 [Source:HGNC Symbol;Acc:HGNC:39832] | 28371 | -0.161 | -0.7276 | Yes |
| 20 | SNX19P2 | sorting nexin 19 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:38115] | 29489 | -0.325 | -0.7007 | Yes |
| 21 | RHOT1P3 | ras homolog family member T1 pseudogene 3 [Source:HGNC Symbol;Acc:HGNC:37839] | 33572 | -1.220 | -0.5966 | Yes |
| 22 | KMT5AP1 | KMT5A pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:30088] | 33749 | -1.282 | -0.3763 | Yes |
| 23 | TERF1P5 | TERF1 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:39686] | 36105 | -2.657 | 0.0266 | Yes |