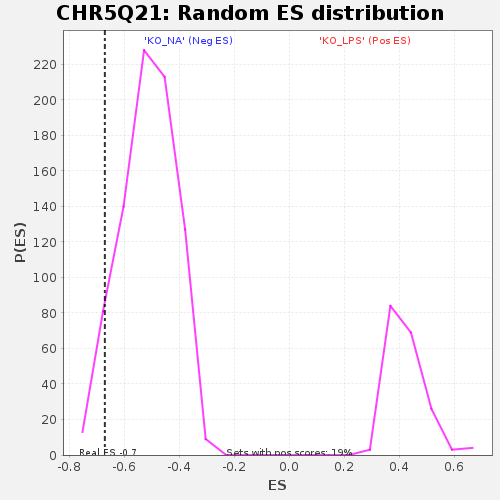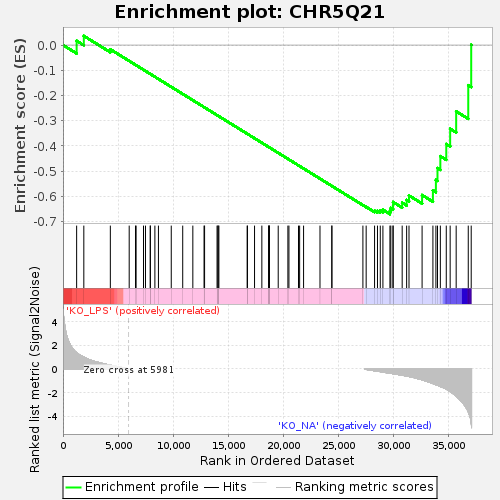
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | CHR5Q21 |
| Enrichment Score (ES) | -0.6705244 |
| Normalized Enrichment Score (NES) | -1.3052158 |
| Nominal p-value | 0.060419235 |
| FDR q-value | 0.6658925 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MACIR | macrophage immunometabolism regulator [Source:HGNC Symbol;Acc:HGNC:25052] | 1238 | 1.359 | 0.0168 | No |
| 2 | CHD1 | chromodomain helicase DNA binding protein 1 [Source:HGNC Symbol;Acc:HGNC:1915] | 1890 | 1.001 | 0.0362 | No |
| 3 | GJA1P1 | gap junction protein alpha 1 pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:4275] | 4297 | 0.319 | -0.0170 | No |
| 4 | RNU6-334P | RNA, U6 small nuclear 334, pseudogene [Source:HGNC Symbol;Acc:HGNC:47297] | 6009 | 0.000 | -0.0632 | No |
| 5 | SLCO6A1 | solute carrier organic anion transporter family member 6A1 [Source:HGNC Symbol;Acc:HGNC:23613] | 6581 | 0.000 | -0.0786 | No |
| 6 | PDZPH1P | PDZ and pleckstrin homology domains 1, pseudogene [Source:HGNC Symbol;Acc:HGNC:51488] | 6636 | 0.000 | -0.0800 | No |
| 7 | SLCO4C1 | solute carrier organic anion transporter family member 4C1 [Source:HGNC Symbol;Acc:HGNC:23612] | 7308 | 0.000 | -0.0982 | No |
| 8 | MTND6P22 | MT-ND6 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:51978] | 7487 | 0.000 | -0.1030 | No |
| 9 | MTATP6P2 | MT-ATP6 pseudogene 2 [Source:HGNC Symbol;Acc:HGNC:44576] | 7924 | 0.000 | -0.1147 | No |
| 10 | AC114324.1 | putative POM121-like protein 1-like [Source:NCBI gene (formerly Entrezgene);Acc:100652833] | 7927 | 0.000 | -0.1148 | No |
| 11 | MTND4LP5 | MT-ND4L pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:42239] | 8341 | 0.000 | -0.1259 | No |
| 12 | LINC01023 | long intergenic non-protein coding RNA 1023 [Source:HGNC Symbol;Acc:HGNC:49004] | 8664 | 0.000 | -0.1346 | No |
| 13 | RNU1-140P | RNA, U1 small nuclear 140, pseudogene [Source:HGNC Symbol;Acc:HGNC:48482] | 9827 | 0.000 | -0.1660 | No |
| 14 | AC099520.1 | novel transcript | 10868 | 0.000 | -0.1941 | No |
| 15 | RN7SL255P | RNA, 7SL, cytoplasmic 255, pseudogene [Source:HGNC Symbol;Acc:HGNC:46271] | 11786 | 0.000 | -0.2189 | No |
| 16 | AC008467.1 | novel transcript | 12804 | 0.000 | -0.2463 | No |
| 17 | RN7SL802P | RNA, 7SL, cytoplasmic 802, pseudogene [Source:HGNC Symbol;Acc:HGNC:46818] | 12853 | 0.000 | -0.2476 | No |
| 18 | LINC02163 | long intergenic non-protein coding RNA 2163 [Source:HGNC Symbol;Acc:HGNC:53024] | 13985 | 0.000 | -0.2782 | No |
| 19 | LINC02115 | long intergenic non-protein coding RNA 2115 [Source:HGNC Symbol;Acc:HGNC:52970] | 14066 | 0.000 | -0.2803 | No |
| 20 | LINC02113 | long intergenic non-protein coding RNA 2113 [Source:HGNC Symbol;Acc:HGNC:52967] | 14080 | 0.000 | -0.2807 | No |
| 21 | EFNA5 | ephrin A5 [Source:HGNC Symbol;Acc:HGNC:3225] | 14151 | 0.000 | -0.2826 | No |
| 22 | MTCYBP22 | MT-CYB pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:51979] | 16719 | 0.000 | -0.3519 | No |
| 23 | PSMC1P5 | proteasome 26S subunit, ATPase 1 pseudogene 5 [Source:HGNC Symbol;Acc:HGNC:39780] | 16745 | 0.000 | -0.3526 | No |
| 24 | RN7SL782P | RNA, 7SL, cytoplasmic 782, pseudogene [Source:HGNC Symbol;Acc:HGNC:46798] | 17388 | 0.000 | -0.3699 | No |
| 25 | RN7SKP230 | RN7SK pseudogene 230 [Source:HGNC Symbol;Acc:HGNC:45954] | 18051 | 0.000 | -0.3878 | No |
| 26 | RN7SKP68 | RN7SK pseudogene 68 [Source:HGNC Symbol;Acc:HGNC:45792] | 18675 | 0.000 | -0.4046 | No |
| 27 | RN7SKP62 | RN7SK pseudogene 62 [Source:HGNC Symbol;Acc:HGNC:45786] | 18679 | 0.000 | -0.4047 | No |
| 28 | RNA5SP189 | RNA, 5S ribosomal pseudogene 189 [Source:HGNC Symbol;Acc:HGNC:43089] | 18727 | 0.000 | -0.4060 | No |
| 29 | RNA5SP188 | RNA, 5S ribosomal pseudogene 188 [Source:HGNC Symbol;Acc:HGNC:43088] | 18728 | 0.000 | -0.4060 | No |
| 30 | LINC01848 | long intergenic non-protein coding RNA 1848 [Source:HGNC Symbol;Acc:HGNC:52663] | 19537 | 0.000 | -0.4278 | No |
| 31 | RN7SKP122 | RN7SK pseudogene 122 [Source:HGNC Symbol;Acc:HGNC:45846] | 20415 | 0.000 | -0.4515 | No |
| 32 | LINC01950 | long intergenic non-protein coding RNA 1950 [Source:HGNC Symbol;Acc:HGNC:52773] | 20510 | 0.000 | -0.4540 | No |
| 33 | LINC02062 | long intergenic non-protein coding RNA 2062 [Source:HGNC Symbol;Acc:HGNC:52907] | 21378 | 0.000 | -0.4774 | No |
| 34 | MIR548P | microRNA 548p [Source:HGNC Symbol;Acc:HGNC:35351] | 21482 | 0.000 | -0.4802 | No |
| 35 | RNU6-1119P | RNA, U6 small nuclear 1119, pseudogene [Source:HGNC Symbol;Acc:HGNC:48082] | 21839 | 0.000 | -0.4898 | No |
| 36 | KRT18P42 | keratin 18 pseudogene 42 [Source:HGNC Symbol;Acc:HGNC:33412] | 23329 | 0.000 | -0.5300 | No |
| 37 | LINC00491 | long intergenic non-protein coding RNA 491 [Source:HGNC Symbol;Acc:HGNC:43428] | 24406 | 0.000 | -0.5591 | No |
| 38 | LINC00492 | long intergenic non-protein coding RNA 492 [Source:HGNC Symbol;Acc:HGNC:43429] | 24407 | 0.000 | -0.5591 | No |
| 39 | OR7H2P | olfactory receptor family 7 subfamily H member 2 pseudogene [Source:HGNC Symbol;Acc:HGNC:31314] | 27225 | 0.000 | -0.6352 | No |
| 40 | GUSBP8 | GUSB pseudogene 8 [Source:HGNC Symbol;Acc:HGNC:42322] | 27521 | -0.028 | -0.6421 | No |
| 41 | GIN1 | gypsy retrotransposon integrase 1 [Source:HGNC Symbol;Acc:HGNC:25959] | 28284 | -0.147 | -0.6572 | No |
| 42 | RNU6-47P | RNA, U6 small nuclear 47, pseudogene [Source:HGNC Symbol;Acc:HGNC:34291] | 28548 | -0.183 | -0.6576 | Yes |
| 43 | EEF1A1P20 | eukaryotic translation elongation factor 1 alpha 1 pseudogene 20 [Source:HGNC Symbol;Acc:HGNC:37893] | 28802 | -0.218 | -0.6564 | Yes |
| 44 | EIF3KP1 | eukaryotic translation initiation factor 3 subunit K pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:44016] | 29041 | -0.256 | -0.6533 | Yes |
| 45 | PJA2 | praja ring finger ubiquitin ligase 2 [Source:HGNC Symbol;Acc:HGNC:17481] | 29679 | -0.351 | -0.6575 | Yes |
| 46 | MTCO1P22 | MT-CO1 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:52087] | 29759 | -0.364 | -0.6462 | Yes |
| 47 | RAB9BP1 | RAB9B, member RAS oncogene family pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:9793] | 29962 | -0.393 | -0.6372 | Yes |
| 48 | PPIP5K2 | diphosphoinositol pentakisphosphate kinase 2 [Source:HGNC Symbol;Acc:HGNC:29035] | 29965 | -0.393 | -0.6227 | Yes |
| 49 | MTCO3P22 | MT-CO3 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:52125] | 30790 | -0.524 | -0.6256 | Yes |
| 50 | MTND5P10 | MT-ND5 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:42272] | 31195 | -0.595 | -0.6145 | Yes |
| 51 | PGAM5P1 | PGAM family member 5, mitochondrial serine/threonine protein phosphatase pseudogene 1 [Source:HGNC Symbol;Acc:HGNC:42467] | 31410 | -0.634 | -0.5968 | Yes |
| 52 | MAN2A1 | mannosidase alpha class 2A member 1 [Source:HGNC Symbol;Acc:HGNC:6824] | 32598 | -0.916 | -0.5950 | Yes |
| 53 | NUDT12 | nudix hydrolase 12 [Source:HGNC Symbol;Acc:HGNC:18826] | 33580 | -1.223 | -0.5763 | Yes |
| 54 | PAM | peptidylglycine alpha-amidating monooxygenase [Source:HGNC Symbol;Acc:HGNC:8596] | 33852 | -1.320 | -0.5349 | Yes |
| 55 | MTND4P35 | MT-ND4 pseudogene 35 [Source:HGNC Symbol;Acc:HGNC:51949] | 33996 | -1.370 | -0.4881 | Yes |
| 56 | FER | FER tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:3655] | 34255 | -1.469 | -0.4408 | Yes |
| 57 | FBXL17 | F-box and leucine rich repeat protein 17 [Source:HGNC Symbol;Acc:HGNC:13615] | 34793 | -1.693 | -0.3927 | Yes |
| 58 | MTND3P19 | MT-ND3 pseudogene 19 [Source:HGNC Symbol;Acc:HGNC:52081] | 35146 | -1.896 | -0.3322 | Yes |
| 59 | MTCO2P22 | MT-CO2 pseudogene 22 [Source:HGNC Symbol;Acc:HGNC:52151] | 35692 | -2.269 | -0.2630 | Yes |
| 60 | FAM174A | family with sequence similarity 174 member A [Source:HGNC Symbol;Acc:HGNC:24943] | 36789 | -3.605 | -0.1594 | Yes |
| 61 | ST8SIA4 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 [Source:HGNC Symbol;Acc:HGNC:10871] | 37057 | -4.531 | 0.0009 | Yes |