Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MISSIAGLIA_REGULATED_BY_METHYLATION_DN |
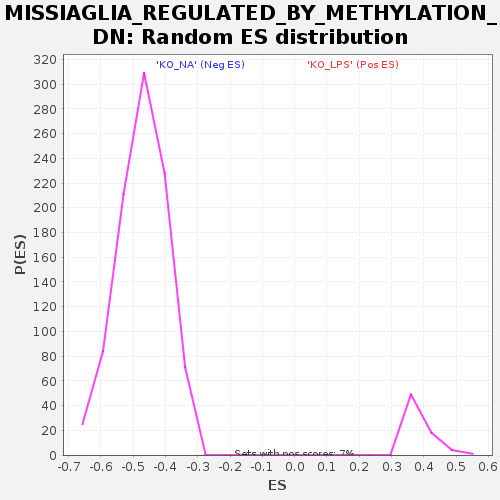
| Enrichment Score (ES) | -0.78525484 |
| Normalized Enrichment Score (NES) | -1.6587437 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0012572696 |
| FWER p-Value | 0.03 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FOSL1 | FOS like 1, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:13718] | 31 | 4.715 | 0.0211 | No |
| 2 | PCDH7 | protocadherin 7 [Source:HGNC Symbol;Acc:HGNC:8659] | 319 | 2.842 | 0.0265 | No |
| 3 | ZNF175 | zinc finger protein 175 [Source:HGNC Symbol;Acc:HGNC:12964] | 1652 | 1.115 | -0.0043 | No |
| 4 | FUT8 | fucosyltransferase 8 [Source:HGNC Symbol;Acc:HGNC:4019] | 1720 | 1.077 | -0.0011 | No |
| 5 | RANBP1 | RAN binding protein 1 [Source:HGNC Symbol;Acc:HGNC:9847] | 2017 | 0.936 | -0.0048 | No |
| 6 | YES1 | YES proto-oncogene 1, Src family tyrosine kinase [Source:HGNC Symbol;Acc:HGNC:12841] | 2186 | 0.862 | -0.0053 | No |
| 7 | AARS1 | alanyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:20] | 2465 | 0.755 | -0.0093 | No |
| 8 | IFT20 | intraflagellar transport 20 [Source:HGNC Symbol;Acc:HGNC:30989] | 2562 | 0.724 | -0.0086 | No |
| 9 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B [Source:HGNC Symbol;Acc:HGNC:5034] | 2574 | 0.721 | -0.0055 | No |
| 10 | SRSF3 | serine and arginine rich splicing factor 3 [Source:HGNC Symbol;Acc:HGNC:10785] | 3238 | 0.542 | -0.0209 | No |
| 11 | FBL | fibrillarin [Source:HGNC Symbol;Acc:HGNC:3599] | 3843 | 0.412 | -0.0354 | No |
| 12 | HMGN2 | high mobility group nucleosomal binding domain 2 [Source:HGNC Symbol;Acc:HGNC:4986] | 4200 | 0.339 | -0.0434 | No |
| 13 | RCC1 | regulator of chromosome condensation 1 [Source:HGNC Symbol;Acc:HGNC:1913] | 4226 | 0.334 | -0.0425 | No |
| 14 | RBMX | RNA binding motif protein X-linked [Source:HGNC Symbol;Acc:HGNC:9910] | 4581 | 0.260 | -0.0509 | No |
| 15 | SNRPA | small nuclear ribonucleoprotein polypeptide A [Source:HGNC Symbol;Acc:HGNC:11151] | 4840 | 0.212 | -0.0569 | No |
| 16 | MDFI | MyoD family inhibitor [Source:HGNC Symbol;Acc:HGNC:6967] | 16090 | 0.000 | -0.3612 | No |
| 17 | RAB26 | RAB26, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:14259] | 17148 | 0.000 | -0.3898 | No |
| 18 | SEMA3A | semaphorin 3A [Source:HGNC Symbol;Acc:HGNC:10723] | 17800 | 0.000 | -0.4074 | No |
| 19 | PIR | pirin [Source:HGNC Symbol;Acc:HGNC:30048] | 19938 | 0.000 | -0.4652 | No |
| 20 | MECOM | MDS1 and EVI1 complex locus [Source:HGNC Symbol;Acc:HGNC:3498] | 22022 | 0.000 | -0.5215 | No |
| 21 | CAV1 | caveolin 1 [Source:HGNC Symbol;Acc:HGNC:1527] | 22238 | 0.000 | -0.5273 | No |
| 22 | CYP2J2 | cytochrome P450 family 2 subfamily J member 2 [Source:HGNC Symbol;Acc:HGNC:2634] | 22613 | 0.000 | -0.5375 | No |
| 23 | NRGN | neurogranin [Source:HGNC Symbol;Acc:HGNC:8000] | 24024 | 0.000 | -0.5756 | No |
| 24 | LINC00667 | long intergenic non-protein coding RNA 667 [Source:HGNC Symbol;Acc:HGNC:27906] | 25121 | 0.000 | -0.6053 | No |
| 25 | ZWINT | ZW10 interacting kinetochore protein [Source:HGNC Symbol;Acc:HGNC:13195] | 26019 | 0.000 | -0.6295 | No |
| 26 | DANCR | differentiation antagonizing non-protein coding RNA [Source:HGNC Symbol;Acc:HGNC:28964] | 26413 | 0.000 | -0.6401 | No |
| 27 | HMGN2P9 | high mobility group nucleosomal binding domain 2 pseudogene 9 [Source:HGNC Symbol;Acc:HGNC:4987] | 26743 | 0.000 | -0.6490 | No |
| 28 | SNRNP40 | small nuclear ribonucleoprotein U5 subunit 40 [Source:HGNC Symbol;Acc:HGNC:30857] | 27374 | -0.000 | -0.6661 | No |
| 29 | ITGB4 | integrin subunit beta 4 [Source:HGNC Symbol;Acc:HGNC:6158] | 27505 | -0.025 | -0.6695 | No |
| 30 | ENOSF1 | enolase superfamily member 1 [Source:HGNC Symbol;Acc:HGNC:30365] | 27845 | -0.082 | -0.6783 | No |
| 31 | DHFR | dihydrofolate reductase [Source:HGNC Symbol;Acc:HGNC:2861] | 29048 | -0.257 | -0.7096 | No |
| 32 | NDUFS3 | NADH:ubiquinone oxidoreductase core subunit S3 [Source:HGNC Symbol;Acc:HGNC:7710] | 29892 | -0.381 | -0.7306 | No |
| 33 | HNRNPL | heterogeneous nuclear ribonucleoprotein L [Source:HGNC Symbol;Acc:HGNC:5045] | 30226 | -0.433 | -0.7376 | No |
| 34 | GMPS | guanine monophosphate synthase [Source:HGNC Symbol;Acc:HGNC:4378] | 30718 | -0.514 | -0.7485 | No |
| 35 | FASN | fatty acid synthase [Source:HGNC Symbol;Acc:HGNC:3594] | 31476 | -0.647 | -0.7660 | No |
| 36 | KIAA0586 | KIAA0586 [Source:HGNC Symbol;Acc:HGNC:19960] | 31532 | -0.657 | -0.7644 | No |
| 37 | ILF3 | interleukin enhancer binding factor 3 [Source:HGNC Symbol;Acc:HGNC:6038] | 31856 | -0.718 | -0.7698 | No |
| 38 | HNRNPA1 | heterogeneous nuclear ribonucleoprotein A1 [Source:HGNC Symbol;Acc:HGNC:5031] | 31982 | -0.750 | -0.7697 | No |
| 39 | CSE1L | chromosome segregation 1 like [Source:HGNC Symbol;Acc:HGNC:2431] | 31993 | -0.752 | -0.7665 | No |
| 40 | HAT1 | histone acetyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:4821] | 32189 | -0.808 | -0.7680 | No |
| 41 | TYMS | thymidylate synthetase [Source:HGNC Symbol;Acc:HGNC:12441] | 32418 | -0.869 | -0.7701 | No |
| 42 | ANP32B | acidic nuclear phosphoprotein 32 family member B [Source:HGNC Symbol;Acc:HGNC:16677] | 32602 | -0.917 | -0.7708 | No |
| 43 | SHMT1 | serine hydroxymethyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:10850] | 32657 | -0.932 | -0.7680 | No |
| 44 | CHEK1 | checkpoint kinase 1 [Source:HGNC Symbol;Acc:HGNC:1925] | 32734 | -0.955 | -0.7656 | No |
| 45 | PCBP2 | poly(rC) binding protein 2 [Source:HGNC Symbol;Acc:HGNC:8648] | 32757 | -0.960 | -0.7617 | No |
| 46 | HMGB1P10 | high mobility group box 1 pseudogene 10 [Source:HGNC Symbol;Acc:HGNC:4994] | 33290 | -1.126 | -0.7709 | No |
| 47 | MICU1 | mitochondrial calcium uptake 1 [Source:HGNC Symbol;Acc:HGNC:1530] | 33823 | -1.311 | -0.7792 | Yes |
| 48 | CAD | carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase [Source:HGNC Symbol;Acc:HGNC:1424] | 33902 | -1.336 | -0.7751 | Yes |
| 49 | DTYMK | deoxythymidylate kinase [Source:HGNC Symbol;Acc:HGNC:3061] | 34013 | -1.377 | -0.7716 | Yes |
| 50 | DUT | deoxyuridine triphosphatase [Source:HGNC Symbol;Acc:HGNC:3078] | 34095 | -1.401 | -0.7673 | Yes |
| 51 | HMGB1 | high mobility group box 1 [Source:HGNC Symbol;Acc:HGNC:4983] | 34140 | -1.424 | -0.7619 | Yes |
| 52 | CDC25C | cell division cycle 25C [Source:HGNC Symbol;Acc:HGNC:1727] | 34146 | -1.427 | -0.7554 | Yes |
| 53 | HDHD5 | haloacid dehalogenase like hydrolase domain containing 5 [Source:HGNC Symbol;Acc:HGNC:1843] | 34159 | -1.431 | -0.7491 | Yes |
| 54 | MTFP1 | mitochondrial fission process 1 [Source:HGNC Symbol;Acc:HGNC:26945] | 34186 | -1.442 | -0.7431 | Yes |
| 55 | HMGB2 | high mobility group box 2 [Source:HGNC Symbol;Acc:HGNC:5000] | 34312 | -1.491 | -0.7395 | Yes |
| 56 | ARHGAP19 | Rho GTPase activating protein 19 [Source:HGNC Symbol;Acc:HGNC:23724] | 34352 | -1.508 | -0.7336 | Yes |
| 57 | CDK1 | cyclin dependent kinase 1 [Source:HGNC Symbol;Acc:HGNC:1722] | 34381 | -1.518 | -0.7273 | Yes |
| 58 | POLD3 | DNA polymerase delta 3, accessory subunit [Source:HGNC Symbol;Acc:HGNC:20932] | 34420 | -1.534 | -0.7212 | Yes |
| 59 | LMNB1 | lamin B1 [Source:HGNC Symbol;Acc:HGNC:6637] | 34439 | -1.540 | -0.7145 | Yes |
| 60 | ALYREF | Aly/REF export factor [Source:HGNC Symbol;Acc:HGNC:19071] | 34441 | -1.541 | -0.7074 | Yes |
| 61 | KNTC1 | kinetochore associated 1 [Source:HGNC Symbol;Acc:HGNC:17255] | 34562 | -1.584 | -0.7032 | Yes |
| 62 | CDC45 | cell division cycle 45 [Source:HGNC Symbol;Acc:HGNC:1739] | 34638 | -1.617 | -0.6978 | Yes |
| 63 | ARL6IP1 | ADP ribosylation factor like GTPase 6 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:697] | 34754 | -1.676 | -0.6931 | Yes |
| 64 | RFC5 | replication factor C subunit 5 [Source:HGNC Symbol;Acc:HGNC:9973] | 34783 | -1.690 | -0.6860 | Yes |
| 65 | PCLAF | PCNA clamp associated factor [Source:HGNC Symbol;Acc:HGNC:28961] | 34837 | -1.712 | -0.6795 | Yes |
| 66 | TRIM28 | tripartite motif containing 28 [Source:HGNC Symbol;Acc:HGNC:16384] | 34840 | -1.714 | -0.6715 | Yes |
| 67 | EXOSC8 | exosome component 8 [Source:HGNC Symbol;Acc:HGNC:17035] | 34907 | -1.757 | -0.6652 | Yes |
| 68 | SMC1A | structural maintenance of chromosomes 1A [Source:HGNC Symbol;Acc:HGNC:11111] | 34943 | -1.772 | -0.6579 | Yes |
| 69 | H2AZ1 | H2A.Z variant histone 1 [Source:HGNC Symbol;Acc:HGNC:4741] | 34958 | -1.777 | -0.6500 | Yes |
| 70 | POLD2 | DNA polymerase delta 2, accessory subunit [Source:HGNC Symbol;Acc:HGNC:9176] | 34987 | -1.797 | -0.6424 | Yes |
| 71 | PRIM1 | DNA primase subunit 1 [Source:HGNC Symbol;Acc:HGNC:9369] | 35085 | -1.852 | -0.6364 | Yes |
| 72 | CDT1 | chromatin licensing and DNA replication factor 1 [Source:HGNC Symbol;Acc:HGNC:24576] | 35143 | -1.895 | -0.6291 | Yes |
| 73 | DCLRE1A | DNA cross-link repair 1A [Source:HGNC Symbol;Acc:HGNC:17660] | 35144 | -1.895 | -0.6203 | Yes |
| 74 | DEK | DEK proto-oncogene [Source:HGNC Symbol;Acc:HGNC:2768] | 35153 | -1.902 | -0.6117 | Yes |
| 75 | CDC6 | cell division cycle 6 [Source:HGNC Symbol;Acc:HGNC:1744] | 35199 | -1.931 | -0.6040 | Yes |
| 76 | LANCL1 | LanC like 1 [Source:HGNC Symbol;Acc:HGNC:6508] | 35333 | -2.016 | -0.5982 | Yes |
| 77 | RPA3 | replication protein A3 [Source:HGNC Symbol;Acc:HGNC:10291] | 35348 | -2.023 | -0.5892 | Yes |
| 78 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35380 | -2.050 | -0.5805 | Yes |
| 79 | RNASEH2A | ribonuclease H2 subunit A [Source:HGNC Symbol;Acc:HGNC:18518] | 35592 | -2.193 | -0.5760 | Yes |
| 80 | CENPM | centromere protein M [Source:HGNC Symbol;Acc:HGNC:18352] | 35632 | -2.222 | -0.5667 | Yes |
| 81 | SMC2 | structural maintenance of chromosomes 2 [Source:HGNC Symbol;Acc:HGNC:14011] | 35759 | -2.331 | -0.5593 | Yes |
| 82 | GMNN | geminin DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:17493] | 35779 | -2.351 | -0.5489 | Yes |
| 83 | MCM5 | minichromosome maintenance complex component 5 [Source:HGNC Symbol;Acc:HGNC:6948] | 35807 | -2.374 | -0.5386 | Yes |
| 84 | CDKN3 | cyclin dependent kinase inhibitor 3 [Source:HGNC Symbol;Acc:HGNC:1791] | 35813 | -2.385 | -0.5276 | Yes |
| 85 | PRKDC | protein kinase, DNA-activated, catalytic subunit [Source:HGNC Symbol;Acc:HGNC:9413] | 35825 | -2.397 | -0.5168 | Yes |
| 86 | RFC4 | replication factor C subunit 4 [Source:HGNC Symbol;Acc:HGNC:9972] | 35830 | -2.402 | -0.5057 | Yes |
| 87 | DLGAP5 | DLG associated protein 5 [Source:HGNC Symbol;Acc:HGNC:16864] | 35981 | -2.530 | -0.4980 | Yes |
| 88 | EZH2 | enhancer of zeste 2 polycomb repressive complex 2 subunit [Source:HGNC Symbol;Acc:HGNC:3527] | 36065 | -2.624 | -0.4881 | Yes |
| 89 | MKI67 | marker of proliferation Ki-67 [Source:HGNC Symbol;Acc:HGNC:7107] | 36125 | -2.680 | -0.4772 | Yes |
| 90 | CENPE | centromere protein E [Source:HGNC Symbol;Acc:HGNC:1856] | 36138 | -2.694 | -0.4650 | Yes |
| 91 | ESPL1 | extra spindle pole bodies like 1, separase [Source:HGNC Symbol;Acc:HGNC:16856] | 36139 | -2.694 | -0.4525 | Yes |
| 92 | SMC4 | structural maintenance of chromosomes 4 [Source:HGNC Symbol;Acc:HGNC:14013] | 36199 | -2.753 | -0.4413 | Yes |
| 93 | BUB1B | BUB1 mitotic checkpoint serine/threonine kinase B [Source:HGNC Symbol;Acc:HGNC:1149] | 36239 | -2.787 | -0.4294 | Yes |
| 94 | STMN1 | stathmin 1 [Source:HGNC Symbol;Acc:HGNC:6510] | 36260 | -2.803 | -0.4169 | Yes |
| 95 | KIFC1 | kinesin family member C1 [Source:HGNC Symbol;Acc:HGNC:6389] | 36274 | -2.821 | -0.4042 | Yes |
| 96 | PSIP1 | PC4 and SFRS1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:9527] | 36305 | -2.851 | -0.3917 | Yes |
| 97 | EBP | EBP cholestenol delta-isomerase [Source:HGNC Symbol;Acc:HGNC:3133] | 36330 | -2.895 | -0.3789 | Yes |
| 98 | ACAT2 | acetyl-CoA acetyltransferase 2 [Source:HGNC Symbol;Acc:HGNC:94] | 36355 | -2.927 | -0.3660 | Yes |
| 99 | RRM1 | ribonucleotide reductase catalytic subunit M1 [Source:HGNC Symbol;Acc:HGNC:10451] | 36359 | -2.931 | -0.3524 | Yes |
| 100 | NCAPD3 | non-SMC condensin II complex subunit D3 [Source:HGNC Symbol;Acc:HGNC:28952] | 36382 | -2.965 | -0.3392 | Yes |
| 101 | CENPF | centromere protein F [Source:HGNC Symbol;Acc:HGNC:1857] | 36421 | -3.011 | -0.3263 | Yes |
| 102 | PAICS | phosphoribosylaminoimidazole carboxylase and phosphoribosylaminoimidazolesuccinocarboxamide synthase [Source:HGNC Symbol;Acc:HGNC:8587] | 36469 | -3.069 | -0.3133 | Yes |
| 103 | UBE2C | ubiquitin conjugating enzyme E2 C [Source:HGNC Symbol;Acc:HGNC:15937] | 36476 | -3.083 | -0.2991 | Yes |
| 104 | MSH6 | mutS homolog 6 [Source:HGNC Symbol;Acc:HGNC:7329] | 36542 | -3.181 | -0.2861 | Yes |
| 105 | NCAPH | non-SMC condensin I complex subunit H [Source:HGNC Symbol;Acc:HGNC:1112] | 36546 | -3.187 | -0.2714 | Yes |
| 106 | LBR | lamin B receptor [Source:HGNC Symbol;Acc:HGNC:6518] | 36547 | -3.188 | -0.2565 | Yes |
| 107 | LIG1 | DNA ligase 1 [Source:HGNC Symbol;Acc:HGNC:6598] | 36576 | -3.229 | -0.2423 | Yes |
| 108 | KIF23 | kinesin family member 23 [Source:HGNC Symbol;Acc:HGNC:6392] | 36599 | -3.262 | -0.2277 | Yes |
| 109 | TMPO | thymopoietin [Source:HGNC Symbol;Acc:HGNC:11875] | 36629 | -3.321 | -0.2131 | Yes |
| 110 | DNMT1 | DNA methyltransferase 1 [Source:HGNC Symbol;Acc:HGNC:2976] | 36635 | -3.330 | -0.1977 | Yes |
| 111 | CCNA2 | cyclin A2 [Source:HGNC Symbol;Acc:HGNC:1578] | 36660 | -3.369 | -0.1827 | Yes |
| 112 | KIF22 | kinesin family member 22 [Source:HGNC Symbol;Acc:HGNC:6391] | 36700 | -3.426 | -0.1679 | Yes |
| 113 | MDC1 | mediator of DNA damage checkpoint 1 [Source:HGNC Symbol;Acc:HGNC:21163] | 36726 | -3.465 | -0.1524 | Yes |
| 114 | HMGB3 | high mobility group box 3 [Source:HGNC Symbol;Acc:HGNC:5004] | 36784 | -3.594 | -0.1373 | Yes |
| 115 | NUSAP1 | nucleolar and spindle associated protein 1 [Source:HGNC Symbol;Acc:HGNC:18538] | 36802 | -3.632 | -0.1208 | Yes |
| 116 | KIF14 | kinesin family member 14 [Source:HGNC Symbol;Acc:HGNC:19181] | 36808 | -3.649 | -0.1040 | Yes |
| 117 | PRC1 | protein regulator of cytokinesis 1 [Source:HGNC Symbol;Acc:HGNC:9341] | 36817 | -3.672 | -0.0872 | Yes |
| 118 | FOXM1 | forkhead box M1 [Source:HGNC Symbol;Acc:HGNC:3818] | 36828 | -3.703 | -0.0702 | Yes |
| 119 | NEK2 | NIMA related kinase 2 [Source:HGNC Symbol;Acc:HGNC:7745] | 36841 | -3.749 | -0.0531 | Yes |
| 120 | H2AZ2 | H2A.Z variant histone 2 [Source:HGNC Symbol;Acc:HGNC:20664] | 36969 | -4.093 | -0.0376 | Yes |
| 121 | CDKN2C | cyclin dependent kinase inhibitor 2C [Source:HGNC Symbol;Acc:HGNC:1789] | 36976 | -4.107 | -0.0186 | Yes |
| 122 | TK1 | thymidine kinase 1 [Source:HGNC Symbol;Acc:HGNC:11830] | 37070 | -4.666 | 0.0005 | Yes |