Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
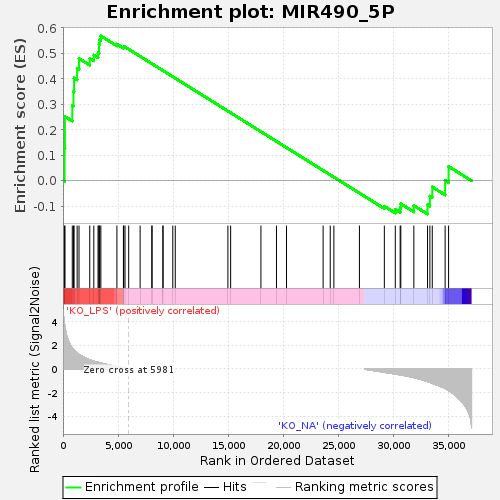
| Upregulated in class | KO_LPS |
| GeneSet | MIR490_5P |
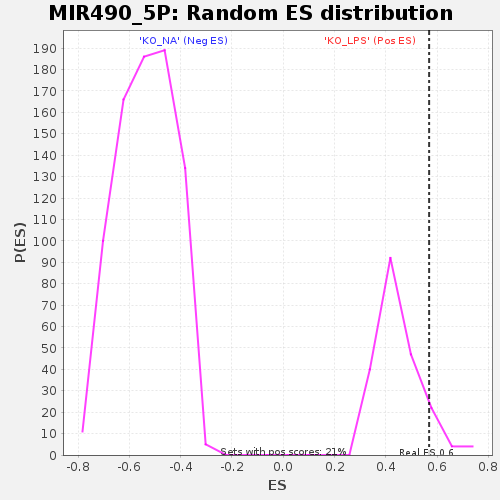
| Enrichment Score (ES) | 0.5689066 |
| Normalized Enrichment Score (NES) | 1.2643516 |
| Nominal p-value | 0.08133971 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | UNC80 | unc-80 homolog, NALCN channel complex subunit [Source:HGNC Symbol;Acc:HGNC:26582] | 144 | 3.781 | 0.1281 | Yes |
| 2 | GCH1 | GTP cyclohydrolase 1 [Source:HGNC Symbol;Acc:HGNC:4193] | 176 | 3.589 | 0.2525 | Yes |
| 3 | ALDH1A2 | aldehyde dehydrogenase 1 family member A2 [Source:HGNC Symbol;Acc:HGNC:15472] | 842 | 1.767 | 0.2963 | Yes |
| 4 | GPALPP1 | GPALPP motifs containing 1 [Source:HGNC Symbol;Acc:HGNC:20298] | 950 | 1.630 | 0.3503 | Yes |
| 5 | WDR44 | WD repeat domain 44 [Source:HGNC Symbol;Acc:HGNC:30512] | 1005 | 1.572 | 0.4037 | Yes |
| 6 | TAOK1 | TAO kinase 1 [Source:HGNC Symbol;Acc:HGNC:29259] | 1284 | 1.322 | 0.4423 | Yes |
| 7 | ARHGAP26 | Rho GTPase activating protein 26 [Source:HGNC Symbol;Acc:HGNC:17073] | 1453 | 1.218 | 0.4803 | Yes |
| 8 | P4HA1 | prolyl 4-hydroxylase subunit alpha 1 [Source:HGNC Symbol;Acc:HGNC:8546] | 2429 | 0.769 | 0.4808 | Yes |
| 9 | SERPINB10 | serpin family B member 10 [Source:HGNC Symbol;Acc:HGNC:8942] | 2797 | 0.650 | 0.4936 | Yes |
| 10 | COA5 | cytochrome c oxidase assembly factor 5 [Source:HGNC Symbol;Acc:HGNC:33848] | 3181 | 0.558 | 0.5028 | Yes |
| 11 | C1orf43 | chromosome 1 open reading frame 43 [Source:HGNC Symbol;Acc:HGNC:29876] | 3268 | 0.535 | 0.5191 | Yes |
| 12 | YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein beta [Source:HGNC Symbol;Acc:HGNC:12849] | 3272 | 0.534 | 0.5377 | Yes |
| 13 | LAMTOR3 | late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 [Source:HGNC Symbol;Acc:HGNC:15606] | 3334 | 0.520 | 0.5542 | Yes |
| 14 | ZEB2 | zinc finger E-box binding homeobox 2 [Source:HGNC Symbol;Acc:HGNC:14881] | 3436 | 0.499 | 0.5689 | Yes |
| 15 | C11orf58 | chromosome 11 open reading frame 58 [Source:HGNC Symbol;Acc:HGNC:16990] | 4893 | 0.201 | 0.5366 | No |
| 16 | USP37 | ubiquitin specific peptidase 37 [Source:HGNC Symbol;Acc:HGNC:20063] | 5483 | 0.097 | 0.5241 | No |
| 17 | ZNF347 | zinc finger protein 347 [Source:HGNC Symbol;Acc:HGNC:16447] | 5514 | 0.088 | 0.5264 | No |
| 18 | ALG10B | ALG10 alpha-1,2-glucosyltransferase B [Source:HGNC Symbol;Acc:HGNC:31088] | 5630 | 0.066 | 0.5255 | No |
| 19 | FOS | Fos proto-oncogene, AP-1 transcription factor subunit [Source:HGNC Symbol;Acc:HGNC:3796] | 5964 | 0.001 | 0.5166 | No |
| 20 | ADCYAP1 | adenylate cyclase activating polypeptide 1 [Source:HGNC Symbol;Acc:HGNC:241] | 7001 | 0.000 | 0.4886 | No |
| 21 | SHISA7 | shisa family member 7 [Source:HGNC Symbol;Acc:HGNC:35409] | 8062 | 0.000 | 0.4600 | No |
| 22 | COLEC11 | collectin subfamily member 11 [Source:HGNC Symbol;Acc:HGNC:17213] | 8087 | 0.000 | 0.4594 | No |
| 23 | CNTN3 | contactin 3 [Source:HGNC Symbol;Acc:HGNC:2173] | 9071 | 0.000 | 0.4328 | No |
| 24 | TTC29 | tetratricopeptide repeat domain 29 [Source:HGNC Symbol;Acc:HGNC:29936] | 9077 | 0.000 | 0.4327 | No |
| 25 | EMCN | endomucin [Source:HGNC Symbol;Acc:HGNC:16041] | 9967 | 0.000 | 0.4087 | No |
| 26 | RTL3 | retrotransposon Gag like 3 [Source:HGNC Symbol;Acc:HGNC:22997] | 10191 | 0.000 | 0.4027 | No |
| 27 | ARHGAP42 | Rho GTPase activating protein 42 [Source:HGNC Symbol;Acc:HGNC:26545] | 14970 | 0.000 | 0.2737 | No |
| 28 | TOX3 | TOX high mobility group box family member 3 [Source:HGNC Symbol;Acc:HGNC:11972] | 15219 | 0.000 | 0.2670 | No |
| 29 | LRATD1 | LRAT domain containing 1 [Source:HGNC Symbol;Acc:HGNC:20743] | 17968 | 0.000 | 0.1928 | No |
| 30 | LMOD1 | leiomodin 1 [Source:HGNC Symbol;Acc:HGNC:6647] | 19378 | 0.000 | 0.1548 | No |
| 31 | INHBC | inhibin subunit beta C [Source:HGNC Symbol;Acc:HGNC:6068] | 20290 | 0.000 | 0.1302 | No |
| 32 | F8A1 | coagulation factor VIII associated 1 [Source:HGNC Symbol;Acc:HGNC:3547] | 23613 | 0.000 | 0.0405 | No |
| 33 | ADGRF3 | adhesion G protein-coupled receptor F3 [Source:HGNC Symbol;Acc:HGNC:18989] | 24260 | 0.000 | 0.0231 | No |
| 34 | CST8 | cystatin 8 [Source:HGNC Symbol;Acc:HGNC:2480] | 24590 | 0.000 | 0.0142 | No |
| 35 | KIRREL1 | kirre like nephrin family adhesion molecule 1 [Source:HGNC Symbol;Acc:HGNC:15734] | 26910 | 0.000 | -0.0484 | No |
| 36 | ARPP19 | cAMP regulated phosphoprotein 19 [Source:HGNC Symbol;Acc:HGNC:16967] | 29171 | -0.276 | -0.0997 | No |
| 37 | HPS5 | HPS5 biogenesis of lysosomal organelles complex 2 subunit 2 [Source:HGNC Symbol;Acc:HGNC:17022] | 30165 | -0.422 | -0.1118 | No |
| 38 | NBR1 | NBR1 autophagy cargo receptor [Source:HGNC Symbol;Acc:HGNC:6746] | 30607 | -0.495 | -0.1064 | No |
| 39 | RANBP10 | RAN binding protein 10 [Source:HGNC Symbol;Acc:HGNC:29285] | 30668 | -0.505 | -0.0904 | No |
| 40 | PKD2 | polycystin 2, transient receptor potential cation channel [Source:HGNC Symbol;Acc:HGNC:9009] | 31849 | -0.716 | -0.0973 | No |
| 41 | BAZ2B | bromodomain adjacent to zinc finger domain 2B [Source:HGNC Symbol;Acc:HGNC:963] | 33092 | -1.060 | -0.0938 | No |
| 42 | CXorf56 | chromosome X open reading frame 56 [Source:HGNC Symbol;Acc:HGNC:26239] | 33305 | -1.127 | -0.0602 | No |
| 43 | APPL1 | adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 1 [Source:HGNC Symbol;Acc:HGNC:24035] | 33522 | -1.199 | -0.0242 | No |
| 44 | SPRED3 | sprouty related EVH1 domain containing 3 [Source:HGNC Symbol;Acc:HGNC:31041] | 34689 | -1.644 | 0.0018 | No |
| 45 | BORA | BORA aurora kinase A activator [Source:HGNC Symbol;Acc:HGNC:24724] | 35003 | -1.805 | 0.0563 | No |