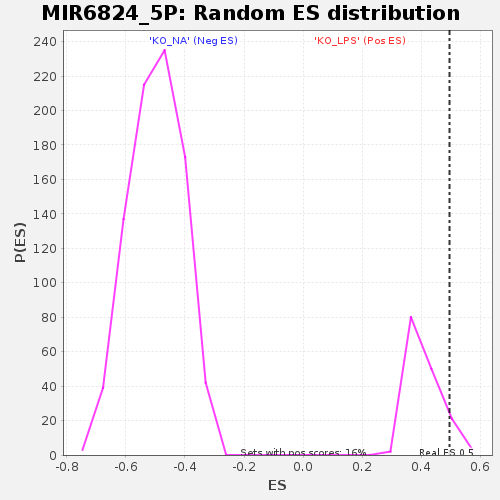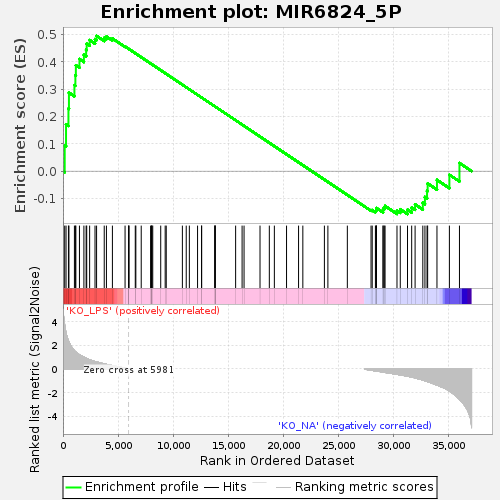
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_LPS |
| GeneSet | MIR6824_5P |
| Enrichment Score (ES) | 0.49552885 |
| Normalized Enrichment Score (NES) | 1.2057508 |
| Nominal p-value | 0.10897436 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | OAS1 | 2'-5'-oligoadenylate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:8086] | 154 | 3.699 | 0.0942 | Yes |
| 2 | OAS2 | 2'-5'-oligoadenylate synthetase 2 [Source:HGNC Symbol;Acc:HGNC:8087] | 276 | 3.036 | 0.1717 | Yes |
| 3 | CLDN12 | claudin 12 [Source:HGNC Symbol;Acc:HGNC:2034] | 494 | 2.381 | 0.2292 | Yes |
| 4 | SLC11A2 | solute carrier family 11 member 2 [Source:HGNC Symbol;Acc:HGNC:10908] | 539 | 2.240 | 0.2876 | Yes |
| 5 | RNF41 | ring finger protein 41 [Source:HGNC Symbol;Acc:HGNC:18401] | 1036 | 1.538 | 0.3151 | Yes |
| 6 | CLIC4 | chloride intracellular channel 4 [Source:HGNC Symbol;Acc:HGNC:13518] | 1124 | 1.448 | 0.3513 | Yes |
| 7 | MAP2K1 | mitogen-activated protein kinase kinase 1 [Source:HGNC Symbol;Acc:HGNC:6840] | 1167 | 1.417 | 0.3879 | Yes |
| 8 | EIF3J | eukaryotic translation initiation factor 3 subunit J [Source:HGNC Symbol;Acc:HGNC:3270] | 1493 | 1.191 | 0.4108 | Yes |
| 9 | NADK | NAD kinase [Source:HGNC Symbol;Acc:HGNC:29831] | 1897 | 0.999 | 0.4264 | Yes |
| 10 | SAR1A | secretion associated Ras related GTPase 1A [Source:HGNC Symbol;Acc:HGNC:10534] | 2099 | 0.899 | 0.4449 | Yes |
| 11 | ZNF800 | zinc finger protein 800 [Source:HGNC Symbol;Acc:HGNC:27267] | 2152 | 0.874 | 0.4668 | Yes |
| 12 | GOLGA7 | golgin A7 [Source:HGNC Symbol;Acc:HGNC:24876] | 2418 | 0.772 | 0.4801 | Yes |
| 13 | UBXN4 | UBX domain protein 4 [Source:HGNC Symbol;Acc:HGNC:14860] | 2906 | 0.623 | 0.4836 | Yes |
| 14 | CD164 | CD164 molecule [Source:HGNC Symbol;Acc:HGNC:1632] | 3048 | 0.593 | 0.4955 | Yes |
| 15 | SLC17A7 | solute carrier family 17 member 7 [Source:HGNC Symbol;Acc:HGNC:16704] | 3743 | 0.432 | 0.4883 | No |
| 16 | ARCN1 | archain 1 [Source:HGNC Symbol;Acc:HGNC:649] | 3942 | 0.390 | 0.4933 | No |
| 17 | ZNF566 | zinc finger protein 566 [Source:HGNC Symbol;Acc:HGNC:25919] | 4485 | 0.282 | 0.4861 | No |
| 18 | KIAA0232 | KIAA0232 [Source:HGNC Symbol;Acc:HGNC:28992] | 5634 | 0.065 | 0.4568 | No |
| 19 | DIS3 | DIS3 homolog, exosome endoribonuclease and 3'-5' exoribonuclease [Source:HGNC Symbol;Acc:HGNC:20604] | 5961 | 0.002 | 0.4481 | No |
| 20 | PCDHA5 | protocadherin alpha 5 [Source:HGNC Symbol;Acc:HGNC:8671] | 5976 | 0.000 | 0.4477 | No |
| 21 | PCDHA4 | protocadherin alpha 4 [Source:HGNC Symbol;Acc:HGNC:8670] | 5977 | 0.000 | 0.4477 | No |
| 22 | GGACT | gamma-glutamylamine cyclotransferase [Source:HGNC Symbol;Acc:HGNC:25100] | 6560 | 0.000 | 0.4320 | No |
| 23 | ANGPTL3 | angiopoietin like 3 [Source:HGNC Symbol;Acc:HGNC:491] | 6613 | 0.000 | 0.4306 | No |
| 24 | EPHA3 | EPH receptor A3 [Source:HGNC Symbol;Acc:HGNC:3387] | 7101 | 0.000 | 0.4174 | No |
| 25 | PCDHAC1 | protocadherin alpha subfamily C, 1 [Source:HGNC Symbol;Acc:HGNC:8676] | 7951 | 0.000 | 0.3945 | No |
| 26 | PROX1 | prospero homeobox 1 [Source:HGNC Symbol;Acc:HGNC:9459] | 8036 | 0.000 | 0.3922 | No |
| 27 | PCDHA13 | protocadherin alpha 13 [Source:HGNC Symbol;Acc:HGNC:8667] | 8058 | 0.000 | 0.3916 | No |
| 28 | PCDHA12 | protocadherin alpha 12 [Source:HGNC Symbol;Acc:HGNC:8666] | 8060 | 0.000 | 0.3916 | No |
| 29 | JPH3 | junctophilin 3 [Source:HGNC Symbol;Acc:HGNC:14203] | 8147 | 0.000 | 0.3893 | No |
| 30 | GRIA4 | glutamate ionotropic receptor AMPA type subunit 4 [Source:HGNC Symbol;Acc:HGNC:4574] | 8871 | 0.000 | 0.3698 | No |
| 31 | PTCHD3 | patched domain containing 3 [Source:HGNC Symbol;Acc:HGNC:24776] | 9275 | 0.000 | 0.3589 | No |
| 32 | AGTR2 | angiotensin II receptor type 2 [Source:HGNC Symbol;Acc:HGNC:338] | 9372 | 0.000 | 0.3563 | No |
| 33 | EPYC | epiphycan [Source:HGNC Symbol;Acc:HGNC:3053] | 10837 | 0.000 | 0.3167 | No |
| 34 | BEND7 | BEN domain containing 7 [Source:HGNC Symbol;Acc:HGNC:23514] | 11187 | 0.000 | 0.3073 | No |
| 35 | TUSC1 | tumor suppressor candidate 1 [Source:HGNC Symbol;Acc:HGNC:31010] | 11470 | 0.000 | 0.2997 | No |
| 36 | G6PC2 | glucose-6-phosphatase catalytic subunit 2 [Source:HGNC Symbol;Acc:HGNC:28906] | 12215 | 0.000 | 0.2796 | No |
| 37 | TYR | tyrosinase [Source:HGNC Symbol;Acc:HGNC:12442] | 12577 | 0.000 | 0.2698 | No |
| 38 | TENM2 | teneurin transmembrane protein 2 [Source:HGNC Symbol;Acc:HGNC:29943] | 12587 | 0.000 | 0.2696 | No |
| 39 | PCDHA3 | protocadherin alpha 3 [Source:HGNC Symbol;Acc:HGNC:8669] | 13799 | 0.000 | 0.2369 | No |
| 40 | PCDHA2 | protocadherin alpha 2 [Source:HGNC Symbol;Acc:HGNC:8668] | 13800 | 0.000 | 0.2369 | No |
| 41 | PCDHA8 | protocadherin alpha 8 [Source:HGNC Symbol;Acc:HGNC:8674] | 13802 | 0.000 | 0.2368 | No |
| 42 | PCDHA6 | protocadherin alpha 6 [Source:HGNC Symbol;Acc:HGNC:8672] | 13803 | 0.000 | 0.2368 | No |
| 43 | CLVS1 | clavesin 1 [Source:HGNC Symbol;Acc:HGNC:23139] | 15671 | 0.000 | 0.1864 | No |
| 44 | ACVR1C | activin A receptor type 1C [Source:HGNC Symbol;Acc:HGNC:18123] | 16258 | 0.000 | 0.1706 | No |
| 45 | ADRA1D | adrenoceptor alpha 1D [Source:HGNC Symbol;Acc:HGNC:280] | 16436 | 0.000 | 0.1658 | No |
| 46 | OPN5 | opsin 5 [Source:HGNC Symbol;Acc:HGNC:19992] | 17885 | 0.000 | 0.1267 | No |
| 47 | ZDBF2 | zinc finger DBF-type containing 2 [Source:HGNC Symbol;Acc:HGNC:29313] | 18730 | 0.000 | 0.1039 | No |
| 48 | NDN | necdin, MAGE family member [Source:HGNC Symbol;Acc:HGNC:7675] | 19187 | 0.000 | 0.0916 | No |
| 49 | COL5A2 | collagen type V alpha 2 chain [Source:HGNC Symbol;Acc:HGNC:2210] | 20293 | 0.000 | 0.0617 | No |
| 50 | ZNF704 | zinc finger protein 704 [Source:HGNC Symbol;Acc:HGNC:32291] | 21384 | 0.000 | 0.0323 | No |
| 51 | INSM2 | INSM transcriptional repressor 2 [Source:HGNC Symbol;Acc:HGNC:17539] | 21772 | 0.000 | 0.0218 | No |
| 52 | KCNIP2 | potassium voltage-gated channel interacting protein 2 [Source:HGNC Symbol;Acc:HGNC:15522] | 23729 | 0.000 | -0.0310 | No |
| 53 | MYO3A | myosin IIIA [Source:HGNC Symbol;Acc:HGNC:7601] | 24046 | 0.000 | -0.0396 | No |
| 54 | SOBP | sine oculis binding protein homolog [Source:HGNC Symbol;Acc:HGNC:29256] | 25812 | 0.000 | -0.0873 | No |
| 55 | CD300E | CD300e molecule [Source:HGNC Symbol;Acc:HGNC:28874] | 27963 | -0.097 | -0.1428 | No |
| 56 | PCDHA10 | protocadherin alpha 10 [Source:HGNC Symbol;Acc:HGNC:8664] | 28071 | -0.114 | -0.1426 | No |
| 57 | P2RY12 | purinergic receptor P2Y12 [Source:HGNC Symbol;Acc:HGNC:18124] | 28361 | -0.158 | -0.1462 | No |
| 58 | PCDHA1 | protocadherin alpha 1 [Source:HGNC Symbol;Acc:HGNC:8663] | 28415 | -0.166 | -0.1433 | No |
| 59 | PCDHA7 | protocadherin alpha 7 [Source:HGNC Symbol;Acc:HGNC:8673] | 28451 | -0.171 | -0.1397 | No |
| 60 | PCDHA11 | protocadherin alpha 11 [Source:HGNC Symbol;Acc:HGNC:8665] | 28457 | -0.172 | -0.1352 | No |
| 61 | ZNF214 | zinc finger protein 214 [Source:HGNC Symbol;Acc:HGNC:13006] | 29050 | -0.257 | -0.1444 | No |
| 62 | RPS6KA3 | ribosomal protein S6 kinase A3 [Source:HGNC Symbol;Acc:HGNC:10432] | 29063 | -0.259 | -0.1378 | No |
| 63 | GK5 | glycerol kinase 5 [Source:HGNC Symbol;Acc:HGNC:28635] | 29166 | -0.275 | -0.1332 | No |
| 64 | CYP27B1 | cytochrome P450 family 27 subfamily B member 1 [Source:HGNC Symbol;Acc:HGNC:2606] | 29236 | -0.287 | -0.1275 | No |
| 65 | BMPR1B | bone morphogenetic protein receptor type 1B [Source:HGNC Symbol;Acc:HGNC:1077] | 30319 | -0.448 | -0.1448 | No |
| 66 | NAA50 | N-alpha-acetyltransferase 50, NatE catalytic subunit [Source:HGNC Symbol;Acc:HGNC:29533] | 30617 | -0.497 | -0.1396 | No |
| 67 | PCDHAC2 | protocadherin alpha subfamily C, 2 [Source:HGNC Symbol;Acc:HGNC:8677] | 31274 | -0.606 | -0.1412 | No |
| 68 | CCSER2 | coiled-coil serine rich protein 2 [Source:HGNC Symbol;Acc:HGNC:29197] | 31653 | -0.680 | -0.1333 | No |
| 69 | SNX9 | sorting nexin 9 [Source:HGNC Symbol;Acc:HGNC:14973] | 31963 | -0.745 | -0.1218 | No |
| 70 | ARL6IP6 | ADP ribosylation factor like GTPase 6 interacting protein 6 [Source:HGNC Symbol;Acc:HGNC:24048] | 32664 | -0.933 | -0.1159 | No |
| 71 | STAU2 | staufen double-stranded RNA binding protein 2 [Source:HGNC Symbol;Acc:HGNC:11371] | 32856 | -0.992 | -0.0947 | No |
| 72 | GSTM4 | glutathione S-transferase mu 4 [Source:HGNC Symbol;Acc:HGNC:4636] | 33048 | -1.047 | -0.0720 | No |
| 73 | CADPS | calcium dependent secretion activator [Source:HGNC Symbol;Acc:HGNC:1426] | 33103 | -1.063 | -0.0451 | No |
| 74 | PIP4P1 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase 1 [Source:HGNC Symbol;Acc:HGNC:19299] | 33948 | -1.352 | -0.0320 | No |
| 75 | MDM1 | Mdm1 nuclear protein [Source:HGNC Symbol;Acc:HGNC:29917] | 35076 | -1.849 | -0.0132 | No |
| 76 | G2E3 | G2/M-phase specific E3 ubiquitin protein ligase [Source:HGNC Symbol;Acc:HGNC:20338] | 35990 | -2.542 | 0.0297 | No |