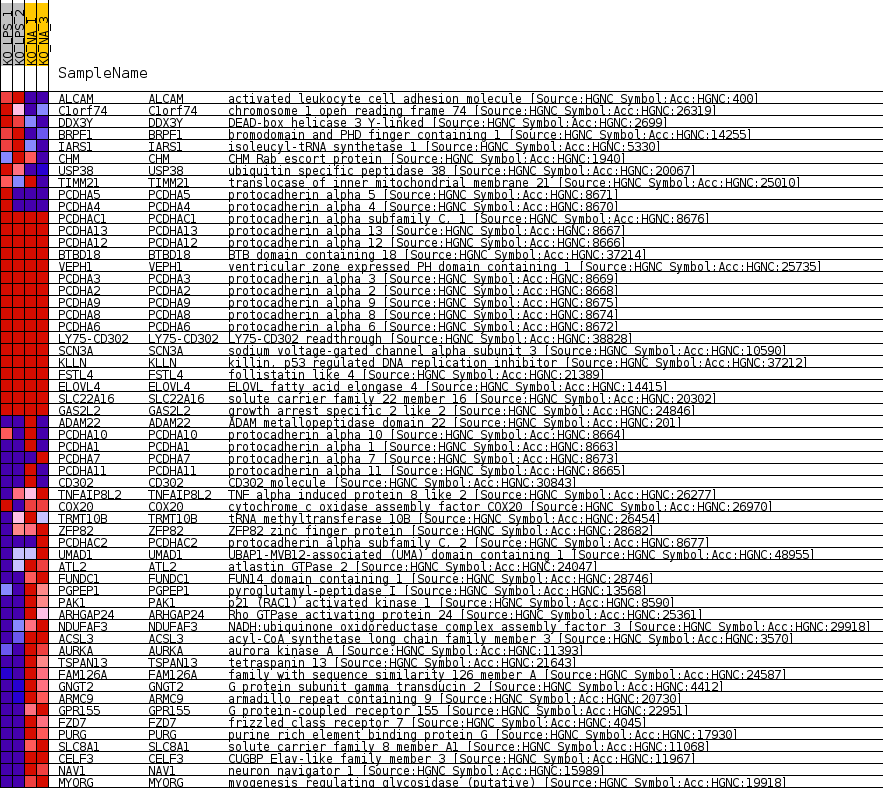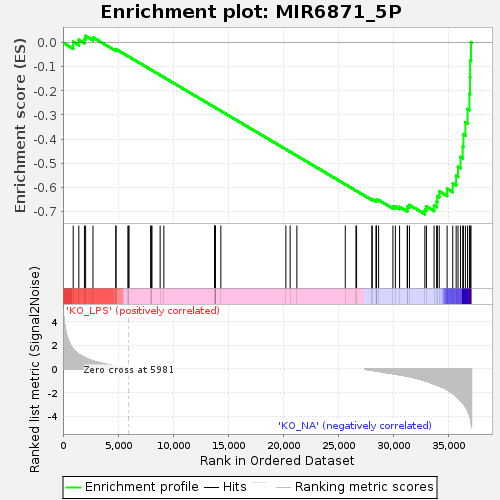
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | greenberg_collapsed_to_symbols.Nof1.cls #KO_LPS_versus_KO_NA.Nof1.cls #KO_LPS_versus_KO_NA_repos |
| Phenotype | Nof1.cls#KO_LPS_versus_KO_NA_repos |
| Upregulated in class | KO_NA |
| GeneSet | MIR6871_5P |
| Enrichment Score (ES) | -0.71165085 |
| Normalized Enrichment Score (NES) | -1.3902735 |
| Nominal p-value | 0.016990291 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ALCAM | activated leukocyte cell adhesion molecule [Source:HGNC Symbol;Acc:HGNC:400] | 926 | 1.658 | 0.0040 | No |
| 2 | C1orf74 | chromosome 1 open reading frame 74 [Source:HGNC Symbol;Acc:HGNC:26319] | 1436 | 1.226 | 0.0117 | No |
| 3 | DDX3Y | DEAD-box helicase 3 Y-linked [Source:HGNC Symbol;Acc:HGNC:2699] | 1945 | 0.971 | 0.0149 | No |
| 4 | BRPF1 | bromodomain and PHD finger containing 1 [Source:HGNC Symbol;Acc:HGNC:14255] | 2052 | 0.922 | 0.0282 | No |
| 5 | IARS1 | isoleucyl-tRNA synthetase 1 [Source:HGNC Symbol;Acc:HGNC:5330] | 2721 | 0.670 | 0.0218 | No |
| 6 | CHM | CHM Rab escort protein [Source:HGNC Symbol;Acc:HGNC:1940] | 4795 | 0.220 | -0.0303 | No |
| 7 | USP38 | ubiquitin specific peptidase 38 [Source:HGNC Symbol;Acc:HGNC:20067] | 4815 | 0.217 | -0.0270 | No |
| 8 | TIMM21 | translocase of inner mitochondrial membrane 21 [Source:HGNC Symbol;Acc:HGNC:25010] | 5890 | 0.013 | -0.0558 | No |
| 9 | PCDHA5 | protocadherin alpha 5 [Source:HGNC Symbol;Acc:HGNC:8671] | 5976 | 0.000 | -0.0581 | No |
| 10 | PCDHA4 | protocadherin alpha 4 [Source:HGNC Symbol;Acc:HGNC:8670] | 5977 | 0.000 | -0.0581 | No |
| 11 | PCDHAC1 | protocadherin alpha subfamily C, 1 [Source:HGNC Symbol;Acc:HGNC:8676] | 7951 | 0.000 | -0.1114 | No |
| 12 | PCDHA13 | protocadherin alpha 13 [Source:HGNC Symbol;Acc:HGNC:8667] | 8058 | 0.000 | -0.1142 | No |
| 13 | PCDHA12 | protocadherin alpha 12 [Source:HGNC Symbol;Acc:HGNC:8666] | 8060 | 0.000 | -0.1142 | No |
| 14 | BTBD18 | BTB domain containing 18 [Source:HGNC Symbol;Acc:HGNC:37214] | 8818 | 0.000 | -0.1347 | No |
| 15 | VEPH1 | ventricular zone expressed PH domain containing 1 [Source:HGNC Symbol;Acc:HGNC:25735] | 9155 | 0.000 | -0.1438 | No |
| 16 | PCDHA3 | protocadherin alpha 3 [Source:HGNC Symbol;Acc:HGNC:8669] | 13799 | 0.000 | -0.2691 | No |
| 17 | PCDHA2 | protocadherin alpha 2 [Source:HGNC Symbol;Acc:HGNC:8668] | 13800 | 0.000 | -0.2691 | No |
| 18 | PCDHA9 | protocadherin alpha 9 [Source:HGNC Symbol;Acc:HGNC:8675] | 13801 | 0.000 | -0.2691 | No |
| 19 | PCDHA8 | protocadherin alpha 8 [Source:HGNC Symbol;Acc:HGNC:8674] | 13802 | 0.000 | -0.2691 | No |
| 20 | PCDHA6 | protocadherin alpha 6 [Source:HGNC Symbol;Acc:HGNC:8672] | 13803 | 0.000 | -0.2691 | No |
| 21 | LY75-CD302 | LY75-CD302 readthrough [Source:HGNC Symbol;Acc:HGNC:38828] | 14330 | 0.000 | -0.2833 | No |
| 22 | SCN3A | sodium voltage-gated channel alpha subunit 3 [Source:HGNC Symbol;Acc:HGNC:10590] | 20232 | 0.000 | -0.4427 | No |
| 23 | KLLN | killin, p53 regulated DNA replication inhibitor [Source:HGNC Symbol;Acc:HGNC:37212] | 20619 | 0.000 | -0.4531 | No |
| 24 | FSTL4 | follistatin like 4 [Source:HGNC Symbol;Acc:HGNC:21389] | 21231 | 0.000 | -0.4696 | No |
| 25 | ELOVL4 | ELOVL fatty acid elongase 4 [Source:HGNC Symbol;Acc:HGNC:14415] | 25624 | 0.000 | -0.5882 | No |
| 26 | SLC22A16 | solute carrier family 22 member 16 [Source:HGNC Symbol;Acc:HGNC:20302] | 26594 | 0.000 | -0.6144 | No |
| 27 | GAS2L2 | growth arrest specific 2 like 2 [Source:HGNC Symbol;Acc:HGNC:24846] | 26649 | 0.000 | -0.6158 | No |
| 28 | ADAM22 | ADAM metallopeptidase domain 22 [Source:HGNC Symbol;Acc:HGNC:201] | 28036 | -0.108 | -0.6514 | No |
| 29 | PCDHA10 | protocadherin alpha 10 [Source:HGNC Symbol;Acc:HGNC:8664] | 28071 | -0.114 | -0.6503 | No |
| 30 | PCDHA1 | protocadherin alpha 1 [Source:HGNC Symbol;Acc:HGNC:8663] | 28415 | -0.166 | -0.6567 | No |
| 31 | PCDHA7 | protocadherin alpha 7 [Source:HGNC Symbol;Acc:HGNC:8673] | 28451 | -0.171 | -0.6546 | No |
| 32 | PCDHA11 | protocadherin alpha 11 [Source:HGNC Symbol;Acc:HGNC:8665] | 28457 | -0.172 | -0.6517 | No |
| 33 | CD302 | CD302 molecule [Source:HGNC Symbol;Acc:HGNC:30843] | 28650 | -0.195 | -0.6535 | No |
| 34 | TNFAIP8L2 | TNF alpha induced protein 8 like 2 [Source:HGNC Symbol;Acc:HGNC:26277] | 29949 | -0.391 | -0.6817 | No |
| 35 | COX20 | cytochrome c oxidase assembly factor COX20 [Source:HGNC Symbol;Acc:HGNC:26970] | 30172 | -0.424 | -0.6803 | No |
| 36 | TRMT10B | tRNA methyltransferase 10B [Source:HGNC Symbol;Acc:HGNC:26454] | 30554 | -0.486 | -0.6821 | No |
| 37 | ZFP82 | ZFP82 zinc finger protein [Source:HGNC Symbol;Acc:HGNC:28682] | 31246 | -0.602 | -0.6903 | No |
| 38 | PCDHAC2 | protocadherin alpha subfamily C, 2 [Source:HGNC Symbol;Acc:HGNC:8677] | 31274 | -0.606 | -0.6804 | No |
| 39 | UMAD1 | UBAP1-MVB12-associated (UMA) domain containing 1 [Source:HGNC Symbol;Acc:HGNC:48955] | 31450 | -0.642 | -0.6739 | No |
| 40 | ATL2 | atlastin GTPase 2 [Source:HGNC Symbol;Acc:HGNC:24047] | 32849 | -0.990 | -0.6943 | Yes |
| 41 | FUNDC1 | FUN14 domain containing 1 [Source:HGNC Symbol;Acc:HGNC:28746] | 32990 | -1.035 | -0.6800 | Yes |
| 42 | PGPEP1 | pyroglutamyl-peptidase I [Source:HGNC Symbol;Acc:HGNC:13568] | 33685 | -1.261 | -0.6767 | Yes |
| 43 | PAK1 | p21 (RAC1) activated kinase 1 [Source:HGNC Symbol;Acc:HGNC:8590] | 33917 | -1.343 | -0.6595 | Yes |
| 44 | ARHGAP24 | Rho GTPase activating protein 24 [Source:HGNC Symbol;Acc:HGNC:25361] | 33984 | -1.366 | -0.6374 | Yes |
| 45 | NDUFAF3 | NADH:ubiquinone oxidoreductase complex assembly factor 3 [Source:HGNC Symbol;Acc:HGNC:29918] | 34155 | -1.431 | -0.6170 | Yes |
| 46 | ACSL3 | acyl-CoA synthetase long chain family member 3 [Source:HGNC Symbol;Acc:HGNC:3570] | 34869 | -1.730 | -0.6060 | Yes |
| 47 | AURKA | aurora kinase A [Source:HGNC Symbol;Acc:HGNC:11393] | 35380 | -2.050 | -0.5839 | Yes |
| 48 | TSPAN13 | tetraspanin 13 [Source:HGNC Symbol;Acc:HGNC:21643] | 35690 | -2.268 | -0.5526 | Yes |
| 49 | FAM126A | family with sequence similarity 126 member A [Source:HGNC Symbol;Acc:HGNC:24587] | 35859 | -2.424 | -0.5148 | Yes |
| 50 | GNGT2 | G protein subunit gamma transducin 2 [Source:HGNC Symbol;Acc:HGNC:4412] | 36086 | -2.639 | -0.4748 | Yes |
| 51 | ARMC9 | armadillo repeat containing 9 [Source:HGNC Symbol;Acc:HGNC:20730] | 36287 | -2.829 | -0.4307 | Yes |
| 52 | GPR155 | G protein-coupled receptor 155 [Source:HGNC Symbol;Acc:HGNC:22951] | 36349 | -2.925 | -0.3812 | Yes |
| 53 | FZD7 | frizzled class receptor 7 [Source:HGNC Symbol;Acc:HGNC:4045] | 36530 | -3.160 | -0.3308 | Yes |
| 54 | PURG | purine rich element binding protein G [Source:HGNC Symbol;Acc:HGNC:17930] | 36725 | -3.461 | -0.2756 | Yes |
| 55 | SLC8A1 | solute carrier family 8 member A1 [Source:HGNC Symbol;Acc:HGNC:11068] | 36895 | -3.874 | -0.2124 | Yes |
| 56 | CELF3 | CUGBP Elav-like family member 3 [Source:HGNC Symbol;Acc:HGNC:11967] | 36945 | -3.997 | -0.1439 | Yes |
| 57 | NAV1 | neuron navigator 1 [Source:HGNC Symbol;Acc:HGNC:15989] | 36959 | -4.043 | -0.0736 | Yes |
| 58 | MYORG | myogenesis regulating glycosidase (putative) [Source:HGNC Symbol;Acc:HGNC:19918] | 37037 | -4.410 | 0.0014 | Yes |